By Constantinos Sophocleous, Luigi Marini, Ropertos Georgiou, Mohammed Elfarargy, and Kenton McHenry
Introduction
Cultural Heritage research is producing huge amounts of data at an ever-increasing rate and size due to the continuous development of more advanced and detailed data extraction technologies and methods. Such research efforts include the pilot projects undertaken by the Cyprus Institute during the establishment of an advanced imaging center for CH and archaeology as part of the LinkSCEEM-2 Cultural Heritage Work Package. The three pilot projects used cutting-edge visualization technologies such as Refectance Transformation Imaging. They generated scientific digital data of archaeological objects and artifacts, such as cylinder seals, ceramics, jewelry and other museum collections, as well as wall paintings and icons from the world known medieval churches of Cyprus.
In these contexts, the digital documentation, storage and management of the aforementioned data as well as providing easy access to researchers is becoming ever more critical. Flexible database storage, processing, metadata extraction and presentation frameworks are needed in order to satisfy these requirements. The development of such database/data repository systems establishes a long-term solution for accessing and storing various kinds of CH data and metadata. These systems must be flexible in terms of being able to have additional data formats support incorporated over time, thus ensuring their continuous update with the latest technological developments, thus maintaining the CH scientific communities’ interest.
The developed framework will be complementary to repositories such as the Inscriptifact database and online library [1]. Having a specialized resource repository for the needs of specific datasets (such as Inscriptifact) must be coupled with a general digital storage and management framework also capable of handling types of data that are not the focus of Inscriptifact. This collaboration will immediately allow the scientific exploration of newly generated CH datasets by an already established user community.
Medici has been and is being developed to address the above. It is a Web 2.0-based general multimedia content management system capable of semantic content management and service/cloud-based workflows. It supports a broad range of throughput-intensive research techniques and community data management. Medici provides scalable storage and media processing, straightforward user interfaces, social annotation capabilities, preprocessing and previewing, metadata extension and manipulation, provenance support and citable persistent data references [2].
Through Medici, users can upload datasets in a variety of formats, including 3D and RTI formats widely used in cultural heritage research and collections [3][4]. Dataset metadata is obtained by extraction services run as data preprocessors and presented to the user along with metadata input by the community for each dataset. Utilizing these extractors Medici is capable of deriving semantically meaningful features for content-based comparison between datasets (e.g. textual data from handwritten text). Previews of large datasets in a variety of formats are also extracted and viewed to avoid the need of (down) loading the whole content on the user’s system or finding the needed software to examine the contents of a file.
Medici 2.0’s scalability/parallelization, flexibility, and robustness, as well as its overall performance, are improved by decoupling the preprocessor software from the main server (with extractors being allowed to run on different machines in a distributed architecture). A powerful and flexible Resource Description Framework (RDF)-enabled database management system is used for storing datasets. Dataset files can be uploaded using a variety of interfaces. Use of the RDF standard makes the datasets’ metadata open and portable. Medici uses the latest Web technologies to display large images smoothly (Seadragon, zoom.it [5]), 3D graphics (X3DOM [6]), video (FFMPEG[7]) and RTI (SpiderGL[8]) along with a special feature added to the RTI web visualization developed by Bibliotheca Alexandrina (BA) to extract 3D models from RTI files (RTI-to-3D) in order to enhance the possibilities of the technology.
Different deployments of Medici can be hosted by different institutions and parts, thus allowing the hosting party better control of access to resources.
Architecture
A diagram describing the dataset upload, preprocessing and previewing process is displayed in Figure 1.

Figure 1. Dataset upload, preprocessing, and previewing process (click image to enlarge).
The system is composed of the following parts:
Web server
Functions and Architecture
Datasets and files are uploaded to the web server using one of an array of methods. One method uses regular Hypertext Markup Language (HTML) forms (both when creating a new dataset to upload its first file as well when viewing a dataset to add files to the dataset). However, files can also be uploaded in other ways, including uploading individual files that do not belong to a dataset.
The web server is responsible for sending a request to the database to store the file or dataset and sending a message to the RabbitMQ message broker [9] defined in the server’s configuration. This message will contain the necessary information pertaining to the file or dataset that is needed by the RabbitMQ broker to distribute the file or dataset’s preprocessing jobs to the available extractors/preprocessors, as well as by the extractors/preprocessors themselves to decide how to process the file or dataset. Datasets contained as files in a zipped archive are inspected by the web server. These files are unzipped to identify the type of dataset whose preprocessing will be handled by extractors on the RabbitMQ bus.
The web server can then passively wait for any auto-extracted file or dataset metadata or previews (generated by the extractors/preprocessors that received a job for the file or dataset) to be uploaded to the server through a representational state transfer (REST) application programming interface (API) [10]. When an upload occurs, the web server calls the database to save the previews/metadata and associate them with the file or dataset.
Extractors can be chained. This means that the resulting file output from an extractor may be fed as input to another extractor, thus allowing even greater flexibility regarding execution of preprocessing subtasks common to the preprocessing of different file or dataset types. For this to happen, an extractor that outputs an intermediate result can upload it as an intermediate result for the file or dataset using the server’s REST API.
This allows scientific preprocessing workflows to be implemented as a preprocessing chain or tree of different extractors. For example (common actual use case), extractor A may convert an RTI (see section “EXTRACTING 3D MODELS FROM RTI”) to an X3D 3D model. Then, that X3D model may be forwarded to extractor B to extract an HTML preview of the model, as well as to a metadata extractor to extract metadata from the 3D model, at the same time. This can generate a preprocessing tree of parallel preprocessing on different extractors. The only information that has to be hardcoded in the extractor itself is that it needs to upload an intermediate result, without needing to know exactly where that result will be forwarded to. The server does not need to know anything either! The intermediate result is forwarded by the server back to the job processing queue, which uses a job key to forward it to the right extractors for further preprocessing (see “Job Broker” section below).
The server then calls the database to save the intermediate result and a new job description is sent to RabbitMQ, now containing the identifiers of both the original file or dataset (with which the final result will be associated) and the intermediate file (which the extractor(s) working on the new job need to download and process). The web server is also responsible for selecting and setting up the previewers that will display the contents of a file or dataset based on the type (i.e. file format) of a file or dataset.
Other functions include working with custom metadata and searches, an interface for adding metadata to a dataset by users based on mapping depending on the institution managing the Medici implementation, searching for datasets having metadata satisfying a query, and text-based search. The server also allows social annotation of datasets (i.e. tagging, notes, comments, custom metadata).
Technologies used
The web server is built on the Play web application framework [11][12], written for allowing coding in Scala and Java. As supported by the framework, the server uses the model-view-controller (MVC) architectural pattern [13]. It relies on a number of plugins for communicating with the RabbitMQ broker, communicating with the database, and authenticating users. It uses dynamic HTML (HTML version 5) webpage generation (views) according to the results of the processing of the input (Scala-based controllers). The models (defined in Scala) are closely associated with collections in the database. Preprocessors and scripts (i.e. previewers) running on users’ browsers communicate with the server using a REST API.
Preprocessors/Extractors
Preprocessors are used for both extracting metadata from datasets and files as well as generating previews for them. Each extractor listens to a RabbitMQ broker via its own named job queue, which is bound to particular routing keys. It is by these keys that RabbitMQ forwards the extraction job messages to the extractors’ queues. Each job message is handled separately. Whenever a job is received, the extractor first downloads the dataset or file having the identifier stated in the job description from the web server via the server’s REST API.
What follows depends on the task each extractor is responsible for. Extractors in general use integrated libraries or external system calls to third-party software to process dataset files to generate the previews or extract the metadata. The third-party software include, for example, 3D model and video processing programs that are installed to the extractor’s environment and are called through Java command-line calls.
Finally, the extractor returns the result back to the web server:
- If the result is an intermediate file for use in extractor chaining, the file is uploaded to the server as an intermediate result to be forwarded to other extractors
through RabbitMQ to continue processing. - If the result is a final preview, it is returned to the server as a preview and then a command is sent to the server to associate the preview with the original dataset or file.
- If the result is a set of dataset or file metadata, a command is sent to the server to associate the metadata with the original dataset or file.
Many extractors of the same type can be run simultaneously on the same or different machines and have the jobs sent by a Medici web server be distributed among them by RabbitMQ as long as they are registered with the same RabbitMQ routing exchange. Also, an extractor may receive messages from more than one Medici web server instance as long as the web servers are registered with the same RabbitMQ routing exchange. This allows for great flexibility, scalability and robustness through a distributed architecture.
Extractors can be implemented using Java or Python.
Previewers
Previewers are used for viewing the contents of a dataset or a file from within the user’s browser. The previewers have been successfully tested on Firefox and Chrome, though with minor adaptations they should work on any modern HTML5-supporting browser.
On the server side, the server selects which previewer to be launched for each preview of each file in the dataset when a dataset page is requested from the web server. The selection is done by comparing each preview’s file type with the file types accepted by each previewer. The latter information is held as JavaScript Object Notation (JSON) data in a public file for each previewer.
Before the previewer scripts are sent to the browser along with the rest of the dataset or file page to be executed, certain variables are set dynamically by the server, mostly for the user’s browser to be able to find the files needed by the previewer on the server (e.g. the dataset files’ URLs).
The previewers use dynamically-added JavaScript scripts downloaded from the web server at runtime and added to the webpage’s DOM (Document Object Model) using jQuery [14]. The previews to be displayed, as well as any other needed files, are also downloaded at runtime using Asynchronous JavaScript and XML (AJAX) calls. After the preview is ready to be displayed, the preview HTML is appended to the webpage’s HTML.
The previewers use browser-side JavaScript and jQuery.
Database
The NoSQL MongoDB is used as the system’s database management system (DBMS). Selecting a NoSQL DBMS offers advantages important for Medici. Flexibility is a key attribute for Medici, as CH community requirements change constantly as described in the Introduction. This gives value to the horizontal scalability of NoSQL. Simplicity of design of NoSQL databases allows the addition of complex features to Medici (such as community-generated dataset metadata) without too much difficulty.
Job broker
If the web server is the heart of Medici 2, the job broker is its veins. The role of the RabbitMQ message broker is to take preprocessing messages from the web server that are sent once a dataset or file is uploaded and distribute them to the extractors that can handle the jobs.
Each extractor, once activated, registers one or more delivery queues on RabbitMQ, on which it listens. Each queue is associated with a particular routing key set, which defines which routing keys a job can have in order for it to be routed to that queue (and thus to the extractor listening to the queue).
Each routing key set is defined by a four-field term like the following:
localhost.file.model.obj.#
In the above, the first field defines the URL or IP of the machine on which the Medici web server implementation sending job messages for the extractors is based. The second field defines whether the extractor takes jobs pertaining to preprocessing a file or a whole dataset. The third and fourth field are for the MIME types of the files (or the types of datasets) that the extractor accepts, and the fifth is the possible subtype of the datasets or files. The syntax of the set definitions uses the conventions specified in the RabbitMQ tutorials.
Metadata
Medici can accept, persist and process two kinds of metadata for each dataset or file:
- Automatically-generated: Generated by metadata extractors and associated with the dataset or file through a REST API call to the web server by the extractors. They usually pertain to the more technical aspects of datasets and files. Their subjects, predicates, and objects can vary from dataset or file type to dataset or file type and from Medici implementation to Medici implementation depending on what metadata are available for each dataset type and dataset, as well as the type of the extractor which extracted the metadata. They cannot be modified by users.
- Community-generated: Generated by the users through completion of an HTML form for each dataset.Their allowed subjects, predicates, and objects are defined on an institutional schema that can differ for each Medici deployment. The schema is formulated by each deployer or institution at its discretion, however it is recommended for the schema to be compatible with the Dublin Core content description model [15] to facilitate metadata sharing between different repositories. The CIDOC Conceptual Reference Model is based on Dublin Core and is specialized for CH metadata and can also be used to facilitate metadata sharing among the CH scientific community [16].
The input metadata, either coming from an extractor or an HTML form, are converted to JSON structures by the extractor or a browser-side script respectively and sent to the web server, where the JSON is parsed back into a binary representation able to be stored in the database.
Metadata is understood by the web server and the JavaScript script manipulating them on the browser-side (for community-generated metadata) as a tree structure (for community-generated metadata) or a list of tree structures (for auto-generated metadata). This distinction applies because different extractors may generate different sets of metadata with no contact points between them for the same dataset or file and so the reasonable option here is to present them as a list. On the other hand, community-generated metadata are generated by users by modifying the whole metadata structure of a dataset using an HTML form at the same time, meaning that they can be merged.

Figure 2. A simple metadata tree for community-generated metadata.
Metadata are formulated as tree structures because each predicate of a metadata subject is either a property of the file or dataset or a subproperty of another property, all describing the same file or dataset. An example of a simple metadata tree (here for community-generated metadata) is presented in Figure 2. The tree structures (one for each dataset) are stored on the web server’s application layer as Scala maps.
Automatically-generated metadata
There are currently metadata extractors for images and RTIs, with more to be added in the future. For a discussion of the metadata extractors for each file type, see the sections discussing Medici’s features for their respective types.
Community-generated metadata
The schema for the community-generated metadata is defined in two comma-separated value files.
- The first file stores the name of each metadata node along with whether the node is a leaf in the metadata tree or a higher-level node. Leaves consist of string data which are input by users and serve as the objects of the description of the dataset, answering the questions generated by the tree paths leading to them. Branches are subproperties of higher-level properties or top-level properties. In the example shown in Figure 2, the leaf node “Title: Ancient Mari” is a leaf answering the question “what is a/the Name of a/the Project Location of a/the Project associated with this dataset?”.
- The second file stores the possible relationships associating nodes with nodes or leaves, along with the cardinalities of each relationship.
The above are automatically enforced by the system when the user adds or modifies a dataset’s community-generated metadata.
Metadata search
Users can search for datasets that contain user-entered metadata that satisfy formulated queries.

Figure 3. Example community-generated metadata search.
The queries are formulated by filling out an HTML form similar to the HTML form for adding/ modifying community-generated metadata. For each level of the metadata tree, the user can select which properties must be satisfied by selected nodes at that level. For example, the query in Figure 3 searches for datasets having a Project, having Cyprus as a Country and a Project Location with “Choirokitia” as a Name, and the Project also having Responsible Institution with a Name, in this case, of “Example Institution.”
Users will be able to use a logical expression-like query formulation to generate queries. For example, he/she may wish to search for datasets satisfying the query above but having Cyprus as a Country OR having a Project Location for which the rest apply, or having Cyprus as a Country and NOT having any Project Locations for which the rest apply.
The JSON-formulated query is sent to the web server, which then parses it as a Scala map, compares it to the community-generated metadata maps of each existing dataset, and returns the list of datasets satisfying the query. The comparison is done using a Depth-First-Search-esque algorithm [17] that recursively checks for all paths in the query tree where there is a path in the dataset’s community-generated metadata tree that satisfies the path. The server formats the result list as HTML and returns it to the client through AJAX to be displayed.
Images
The support of multi-format and multi-resolution image storage and presentation is highly beneficial to CH communities due to the development of image acquisition instruments that satisfy current demands of research analysis in archaeology. Medici currently supports the JPEG/JPG, PNG, TIFF, GIF and BMP image formats.
Moreover, image processing, pyramidal multi-resolution conversion and consequently, optimization of high-resolution images (giga-images), is carried out by the extractor on the virtual server in order to store all necessary image datasets to the appropriate multi-dimensional matrix array and to be delivered to the user when needed thus creating a suitable and efficient way to draw high-resolution web images on screen without affecting the high frame-rate and fast open experience.
Deep Zoom
Viewing of large images efficiently and without loss of quality is achieved by constructing an image pyramid using a technique similar to frustum culling. Specifically, when a large image is to be viewed in a limited screen space, only a part of it can be viewed in full quality and the rest of the information in the image is essentially occluded. Consequently, data transmission size is limited by tiling the image at various scales in a preprocessing step executed server-side so that only the tiles needed for the currently shown scale and position are transmitted. Deep Zoom, a technology developed by Microsoft as part of Microsoft Silverlight [18], works exactly like that- with the user initially seeing a lower-resolution view of the entire image which they can then zoom in to see portions of the image in more detail.
Zoom.it
Zoomit.js (http://zoom.it/h.js) is a JavaScript library also used by the Zoom.it web service. It relies on the technology behind the Seadragon gigaimaging viewer, which utilizes Deep Zoom. This library is used by Medici 2 to view large images that have already been split up into various levels of Deep Zoom tiles (i.e. the image pyramid) by a dedicated image pyramid extractor. The pyramids are loaded on the web server together with an XML Deep Zoom Image file (DZI) containing basic image metadata needed by zoom.it.
Gigaimage processing
When an image is uploaded on the web server, a message is sent to RabbitMQ for any extractors that can process images. If there are any active gigaimaging extractors registered with RabbitMQ, a gigaimaging extractor will receive the job message for the image.
The job message will contain the image’s size. If the image’s size is below a threshold registered value, the job is rejected and no XML file for zoom.it or the image pyramid tiles are generated. Consequently when a user requests to view the dataset containing the image, no zoom.it preview will be found by the web server and therefore the original image will be shown, embedded with an HTML tag.
If the size of the image exceeds the threshold, it is given an identifier and a zoom.it XML file is generated and registered. That identifier will be used to associate the image tiles with the preview and thus with the original image. Then the image tiles for all the image zoom levels are generated, concurrently uploaded back to the web server, and associated with the appropriate preview.
Zoomit.js accesses the image’s pyramid tiles via a virtual directory structure generated on the web server for each preview. This structure is implemented with Scala functions that take the parts of the virtual directory path as parameters and return the requested tile after querying the database where the tiles are stored. These parameters include the tile’s level and 2D-coordinates of its position on the level.
An example of a zoom.it preview can be seen in Figures 4 and 5, where it is used for visualizing ancient cylinder seals and byzantine wall paintings from Cyprus.
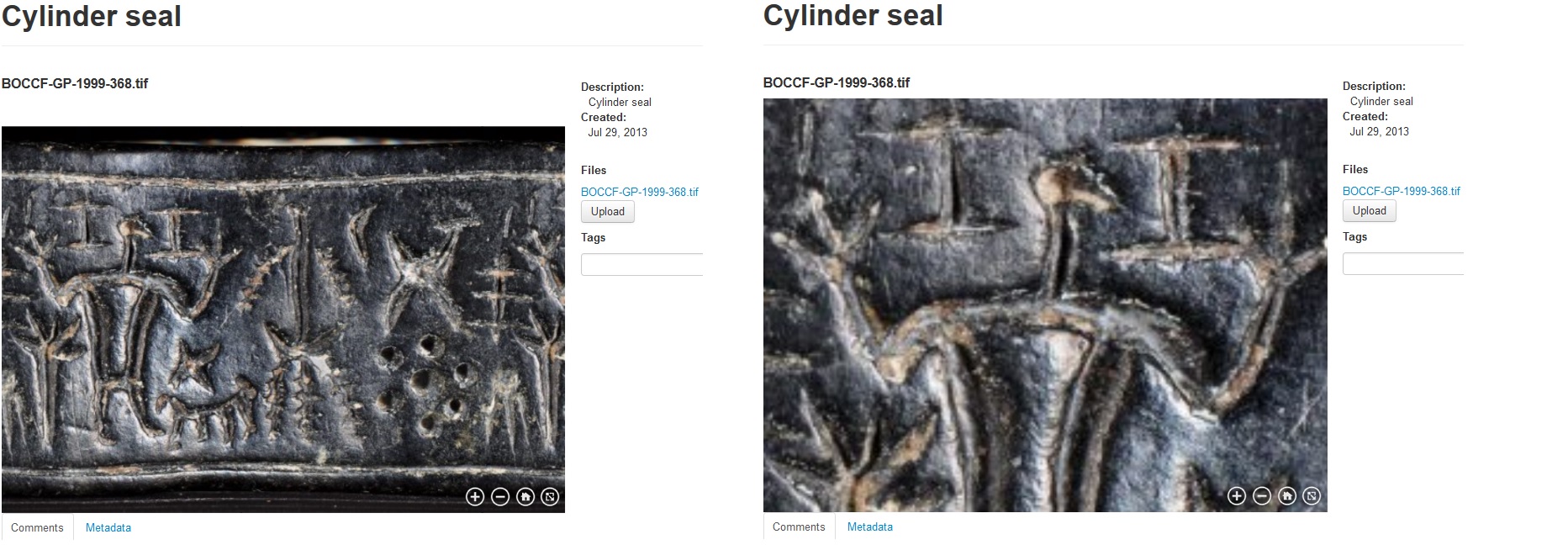
Figure 4. A two-dimensional giga-image of a cylinder seal dating to the 13th century BC taken with a large format camera system and previewed in Medici’s 2.0 web viewer. Courtesy of the Bank of Cyprus Cultural Foundation.
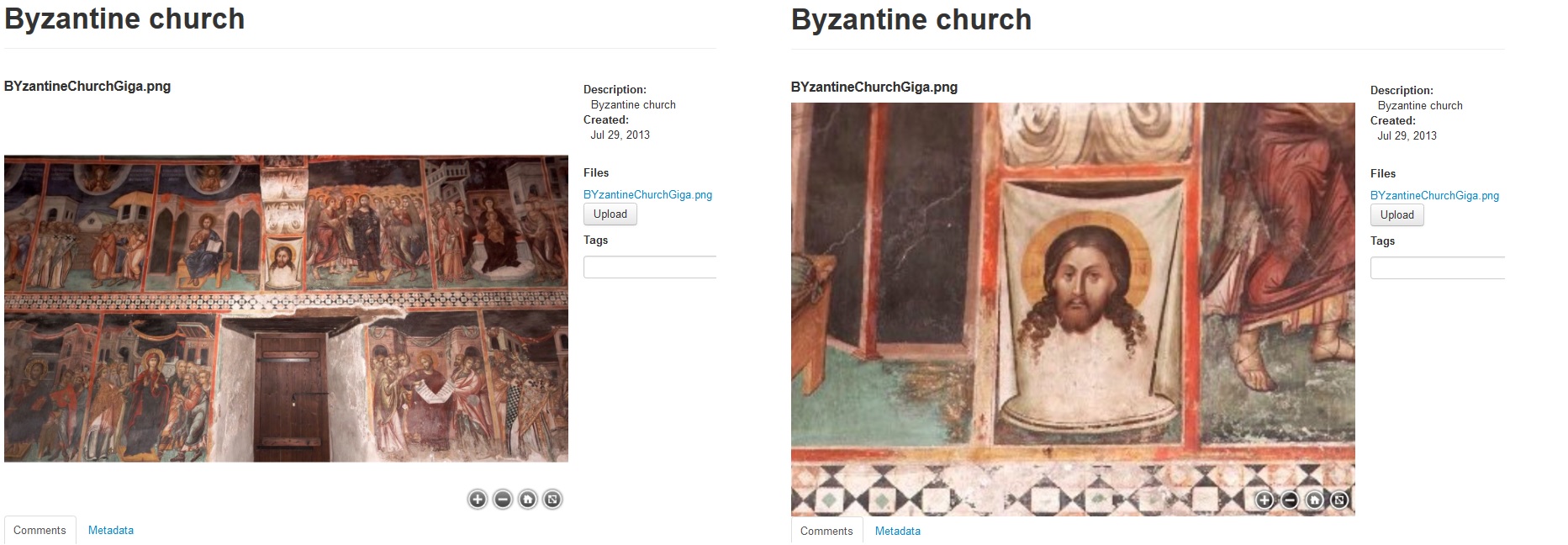
Figure 5. Giga-image of byzantine wall paintings taken inside the monastery of St. John Lambadistis in Cyprus and previewed in the web viewer. Courtesy of the Department of Antiquities in Cyprus.
Image technical metadata
There are currently two different image metadata extractors, either of which can be used at the discretion of the deploying institution, each extracting different sets of metadata. The first uses the ImageMagick image processing suite [19] and the second uses the Java-based image metadata extractor library developed by Drew Noakes [20]. The ImageMagick extractor extracts many more details, even though it needs ImageMagick to be installed on the machine running the ImageMagick extractor.
3D Models
Complete digital documentation in archaeological and Cultural Heritage research is a multidimensional process. New opportunities and challenges for the development of web-based Virtual Reality (VR) applications in these research fields have been the direct consequence of advances in the field of three-dimensional representation and Internet-related technology [21]. High accuracy and multi level of detail (LOD) models can be obtained by using various methods for 3D model creation such as laser scanning technology, photogrammetry and 3D modeling (architectural reconstruction techniques) [22]. Nonetheless, the fusion of these techniques during post-processing may occur, thus leading to the creation of highly complex models with a diversity of information encapsulated within a single file. The development of a web-based application for user access and interactive exploration of three- dimensional models has been studied and worked upon since 1995. The WEB 3D consortium composes open standards for real time 3D data and models exchanged over the Internet. Virtual Reality Modeling Language (VRML) became the first web 3D format [23]. The latest successor of VRML is X3D, providing a flexible solution for real time 3D representation and communication for Medici.
The system architecture of the 3D processing pipeline consists of open source tools and free software, thus providing full transparency on adopted methodology and data processing methods providing a cost effective solution both for server and client. The main feature of this web VR system previewer is to provide the user with a completely new visit experience based on a free interactive exploration interface of the object (i.e., not constrained by any predefined pathway) and the opportunity to get more detailed information on specific parts of interest.
Medici currently supports 3D file formats that are widely used in CH such as Wavefront (obj), Polygon File Format (ply), X3D (x3d) format and 3D models embedded in PDF (3DPDF). Models in the obj or ply format are converted to x3d and then translated to their center of rotation. “Virtual scenes” containing multiple objects can also be previewed in 3D fly-through and walk-through mode (primary mode is “examine” mode). If the model needs optimization for smooth transmission over the internet, it is converted in an adequate LOD, before being converted to the X3DOM format (that is, HTML5 files) and sent back to the server as a preview to be added to the DOM of the 3D previewer and displayed upon request by the user.
There are also previewers for 3DPDF (also used for general PDF viewing). One of them was written by NCSA and uses Mozilla’s PDF.js library [24], while the other is a simple integration of Adobe Acrobat using the associated Acrobat browser plugin.
X3D extraction
The X3D extractor is responsible for converting obj and ply models to X3D and converting them if needed through external calls to MeshLab’s Meshlabserver [25] and setting job identity flags on each of the models uploaded by the client.
For the X3D extractor to work, MeshLab must be installed on the extractor’s machine. The extractor accepts jobs from RabbitMQ for obj, ply and X3D. However, if the 3D model is not in X3D form, MeshLab’s Meshlabserver command-line interface is called by the extractor to generate the equivalent X3D model.
On the other hand, if the model’s file size is above a pre-register boundary size, the client is responsible for calling/activating the extractor program as an input parameter and a dynamically-generated Meshlabserver re-meshing/simplification script is used. A new call to convert the model to the adequate LOD is then generated according to the client’s preset boundaries in order to reduce its complexity and thus its size.
Finally, a flag is set on the job received by the extractor indicating that the first phase of the processing of the model was completed and the post-processed model is uploaded back to the web server as an intermediate result together with the model’s new job flag. The web server then transmits the intermediate result to the HTML5-X3DOM extractor for preparation for front-end display
HTML5-X3DOM extraction
Converting X3D models to X3DOM/HTML5 facilitates and optimizes their integration with the X3D previewer’s HTML DOM structure. This is done by the X3DOM extractor, which uses aopt, a command-line tool bundled with the InstantReality framework [26].
For the HTML5-X3DOM extractor to work, InstantReality must be installed on the extractor’s machine.
Models with separate materials/texture files
In special cases and in order to have obj and X3D files display their geometry with the adequate color information, models at times have separate material/texture files. Medici 2.0 considers this case. Those files can be still previewed correctly if they are uploaded contained in a ZIP file with their corresponding materials/textures. The web server uses a utility function that unzips the file and uses rules regarding the existence of files of certain formats in the zipped dataset to resolve the type of model contained in the ZIP file (whether it is a zipped obj, a zipped X3D, or something different). The type discovered is sent as part of the routing key to RabbitMQ when the file’s extraction job is sent.
After the automated post-processing of the model with its textures from the X3D extractor, the HTML5-X3DOM extractor completes work on the X3D preview and sends back the HTML5 X3DOM as the file’s final preview.
X3DOM previewer
The X3DOM previewer works by dynamically downloading the HTML5 file in which the X3D preview of the model is encapsulated, and embedding it into an X3D HTML5 element. Afterwards, events generated by the new elements activate functions in the X3DOM JavaScript library, initializing the model’s preview through an X3D scene.
The preview allows (among other functions) rotating the model, zooming, panning, displaying model statistics, changing rendering geometry mode from default view to wireframe or vertex view, with or without texture, and surface depth map simulation.
X3dom.js (http://www.x3dom.org/download/dev/x3dom.js accesses any image textures via a virtual directory structure generated on the web server for each preview, which, for files uploaded as ZIP files, simulates the original ZIP file’s internal directory structure. This structure is implemented with Scala functions that take the parts of the virtual directory path as parameters and return the requested image texture after querying the database where the image textures are stored. These parameters include the (virtual) path to the image texture, as it was in the original ZIP file containing the textures of the original model.
3D model preview examples (with the depth maps and model statistics also being displayed) are shown in Figures 6 and 7.

Figure 6. Hellenistic-Roman theater, 300 BC-365 AD, previewed in the web 3D viewer. Courtesy Cyprus Dept. of Antiquities-University of Sydney.

Figure 7. Ancient vessel from the Pyrgos area of Cyprus, previewed in the web viewer. Courtesy of the 3D-coform EU project.
RTI
The application of Reflectance Transformation Imaging Technology (RTI) has offered great possibilities for research as well as for the documentation and preservation of cultural heritage objects and works of art. Polynomial Texture Map (PTM) is a subset of the RTI method and was developed by Hewlett-Packard Imaging Labs at the beginning of the past decade [27][28] and enhanced by the West Semitic Project at University of Southern California (USC) and Cultural Heritage Imaging (CHI). PTM addresses the challenges in the photography of objects’ faded, damaged or badly preserved surfaces especially when they feature inscription, decorative patterns and designs. The surfaces of an array of archaeological objects and works of art such as stone or clay, marble, plaques, coins, paintings, mosaics, sculpture, jewelry and other small objects (see Figure 8), present optimal study cases for the utilization of RTI technology.

Figure 8. An ancient replica coin that shows the capabilities of RTI photography. Self-shadows and interreflections are such derivatives presented on the upper-left corner in comparison with the default mode on the lower-left side that has no such properties.
RTI/PTM viewer
The PTM algorithm synthesizes the data from images taken under varying lighting directions to create a single image that can be examined on a RTI/PTM viewer with a “virtual spotlight”. The viewer allows the user to move the light angle intuitively in real time, so that the combination of light and shadow representing the relief features of the object’s surface can be freely altered. RTI also permits the enhancement of the subject’s surface shape, color and luminance attributes, which allows one to extract detail out of the surface that cannot be otherwise derived (see Figure 9).

Figure 9. This snapshot is taken from the Inscriptifact desktop RTI viewer developed by West Semitic Project at USC where the user is able to interact with the artifact using a “virtual torch” for dynamic illumination and surface enhancement.
Medici currently supports viewing real-time web RTI through a Java applet developed by Clifford Lyon [29] (see Figure 10). There are three stand-alone desktop viewers that can interpret RTI/PTM files but for the web just the viewer mentioned above [30][31][32] is used. The current online RTI viewer has pitfalls concerning incompatibility with current W3C standards.

Figure 10. Replica of an ancient coin from Petra, Jordan, using the current PTM viewer.
In the future, the current viewer will be replaced with one based on the SpiderGL library, which is based on WebGL. This will allow direct execution of the viewer by the user’s browser without the need to use a third-party plugin, improving cohesion between the user’s Graphics Processing Unit, the viewer, and the user’s browser, and also improving security.
Medici 2.0 will currently avoid the use of the RTI Java applet, but instead will use an innovative method for extracting 3D models from RTI files for front-end previewing. Based on this the user can recognize the object and then (down)load the original high-resolution RTI file to his/her desktop for actual interaction.
RTI metadata
RTI technical metadata is extracted by a standalone preprocessor in the same manner as metadata for traditional web images are. The preprocessor downloads the RTI file from the web server and then reads its topmost lines for the types of metadata included in the most common RTI file formats as defined in the PTM file format specification by Hewlett-Packard. The metadata is parsed to JSON and returned to the web server to be associated with the RTI graphic.
Extracting 3D Models From RTI
The ability to interactively change light direction and apply various filters makes RTI one of the best techniques for examining archaeological artifacts. Human perception of highlights and shadows in a 2D image of a surface helps the viewer interpret the surface topology. However, this ability to perceive surface topology varies from person to person. Also, some surfaces with varying contrast and surface characteristics might be difficult to perceive even with dynamic lighting [33].
RTIs capture both color and geometric properties of the object, which makes it possible to use them to reconstruct 3D approximations of the original surfaces. Doing this greatly improves the perception of the artifacts’ surface details. The approach used is based on an approach similar to that of photometric stereo [34] and uses three 2D texture maps that are extracted from the RTI to then reconstruct the final 3D surface given known lighting directions. Luminance info is used to estimate a normal map which is integrated to obtain a height map. The height map is used to build a 3D surface by shifting vertices of a Delaunay mesh. Color info is used to extract a uniformly lit diffuse map that is used for surface texture mapping.
Maps used
Diffuse Map
Diffuse maps (a.k.a. albedo maps) define the main color of the surface. A good diffuse map should contain only color information without any directional light effects, inter-reflections, specular highlights or self-shadowing. Having any of these in a diffuse map means that the object will respond in an incorrect way to virtual incident light; such as casting shadows in the wrong directions or showing highlights where no direct incident light hits the object.
PTM allows the extraction of accurate diffuse maps because the luminance and chromaticity information are stored separately. In LRGB PTM format the color information exists out of the box. For RGB PTM format, similar results can be obtained by casting light perpendicular to surface per texel to make sure all texels get the same amount of light. This results in a uniformly lit texture that is ideal for use as a diffuse map.
Normal Map
A Normal Map is a texture map containing surface normals at each texel stored as RGB. Directional Lighting information for each pixel is already stored in a PTM, which makes it possible to get a very good estimate of the surface normal at that pixel. Since Luminance is maximum when incident light is perpendicular to surface (i.e. incident light vector = surface normal), the surface normal at each texel can be estimated as the orientation maximizing light response.
Height Map
Height maps are grayscale texture maps in which each pixel’s white level corresponds to the height value of a vertice of a 3D grid mesh (usually in the Z direction). Height maps used here are generated by integrating normal maps obtained as described in the previous section. Normals at every surface point are perpendicular to the height map gradient. Integration is not always guaranteed to yield precise results since it is based on an estimated normal map. Also, information about surface discontinuities is lost in normal maps. The 3D models generated are good approximations of the real surface.
Height map generation is implemented in an iterative manner in which each iteration improves contrast between low and high points. Height map pixels are initiated at zero height. On each new iteration, each pixel’s height is slightly modified according to the slopes of the surrounding pixels’ normals and their heights in the current iteration. Contributions from all eight surrounding pixels will be averaged and added to the current height. See Figure 11 for a summary of surrounding pixels, contributions, and associated signs, where Nx and Ny are the X and Y components, respectively, of the surface normal.
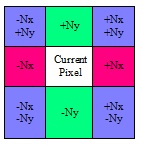
Figure 11. Contributions of surrounding pixels to the current pixel height when generating the height map from the normal map.
Testing the algorithm
In testing done by Bibliotheca Alexandrina IT research center (BA), a 2D Delaunay triangulation was used to generate rectangular grids that were deformed using resulting height maps, and texture-mapped using diffuse and normal maps. The quality of the generated models improved
proportionally with the number of iterations used. Iterations beyond 100 000 iterations had no noticeable effect.
Further testing of the algorithm took place at the Cyprus Institute’s Imaging Cluster for Archaeology and Cultural Heritage (ICACH) [35]. The testing showed that for low and mezzo-relief RTI the algorithm generated 3D models of good quality with no more than 10 000 iterations needed. An example of testing the algorithm is shown in Figure 12.

Figure 12. Example of a PTM file and its 3D model generated by the RTI-to-3D algorithm.
Implementation
The algorithm will be implemented as a standalone command-line program that will take as input the desired number of height map generation iterations, the desired height modifier for the generated 3D model, and the RTI file and output the generated 3D model. This program will be called using a standard extractor taking the above input parameters as
its input. The result of the extraction will be sent to the 3D extractors for further processing, as it is for all 3D models.
Future Directions
Immediate plans
The community-generated metadata search will be enhanced to enable use of logical expression-like query formulation to generate queries with AND, OR, NOT operators. Technical metadata will also be searchable.
Integration of more MeshLab features in Medici will be attempted (if supported by X3DOM). These may include shading modes, a measuring caliber, community-generated annotations on the model display, and being able to change the view’s light source.
Support for more file formats and model types will be added, including printer-ready stereolithography (STL) diagrams, Virtual Reality spherical panoramas, and videos.
Further designs
Medici metadata will be able to be integrated with Inscriptifact and other Cultural Heritage repositories.
Concerning 2D high-resolution imaging, the IIPImage framework [36] will serve as a mediator for flexibility on the presentation of more specialized CH imaging (e.g., able to support multispectral imaging and real time annotation).
Regarding 3D, technical metadata extractors may be added and also the capability to generate 3D models from image datasets produced by photogrammetry similar to the ARC 3D web service [37]. Furthermore, the use of Medici’s interactive system could therefore be potentially extended to more complex virtual exploration such as a digitized archaeological site, to serve not just as a previewer but as an intuitive highly VR environment for web and thus becoming an interactive learning environment, a “virtual world” [38].
A SpiderGL-based viewer will replace the current RTI Java applet once the viewer is developed and made stable by the digital CH communities.
Medici will also be able to extract semantically-meaningful features for content-based comparison (e.g. textual data from handwritten text). This will be made possible by constructing descriptors of each file according to derivations of semantically-meaningful features from a file’s data[39][40][41][42]. The generated descriptors for each file can be compared with descriptors of the same type generated from search query data.
Conclusion
Though still in development, Medici 2 already supports a broad range of throughput-intensive research techniques and community data management. Many file and dataset formats can be uploaded, analyzed and visualized. These include the latest formats used by the CH scientific community, such as large, detailed 3D models and RTI, as well as the ever-present large images and audio/video. Not only can Medici 2 extract technical metadata from files and datasets, it also allows searching of metadata and social metadata generation according to each implementer’s user-input metadata schema.
The above, together with the many important additions scheduled for the future, uniquely sets Medici 2 apart in satisfying the CH scientific community’s dataset management, analysis and visualization needs.
Acknowledgments
The authors thank the Cyprus Institute, the LinkSCEEM-2 project and its partners, the National Centre for Supercomputing Applications (University of Illinois) (NCSA) and Bibliotheca Alexandrina as organizations for providing them the means and guidelines enabling their contributions to the development of Medici. Moreover, they thank every member of staff in CyI, NCSA and Bibliotheca Alexandrina who provided them with user requirements and technical assistance.
Notes
[1] L. Hunt, M. Lundberg, and B. Zuckerman, “InscriptiFact: A virtual archive of ancient inscriptions from the Near East”, in International Journal on Digital Libraries, vol. 5, no. 3, pp. 153-166, May 2005.
[2] L. Marini et al, “Medici: A Scalable Multimedia Environment for Research”, white paper, presented at Microsoft eScience Workshop 2010, Berkeley, CA, Oct. 2010.
[3] The International Committee for Documentation of Cultural Heritage (CIPA). http://cipa.icomos.org
[4] M. Mudge et al, “Image-Based Empirical Information Acquisition, Scientific Reliability, and Long-Term Digital Preservation for the Natural Sciences and Cultural Heritage”, in Eurographics 2008, Hersonissos, Greece, 2008.
[5] Zoom.it http://zoom.it
[6] J. Behr, P. Eschler, Y. Jung, and M. Zöllner, “X3DOM: a DOM-based HTML5/X3D integration model”, in Proc.of the 14th Int. Conf. on 3D Web Technology, Darmstadt, Germany, Jun. 2009, pp. 129-135.
[7] S. Tomar, “Converting video formats with FFmpeg”, in Linux J., vol.2006, no.146, pp. 10, Jun. 2006.
[8] M. Di Benedetto, F. Ponchio, F. Ganovelli, and R. Scopigno, “SpiderGL: a JavaScript 3D graphics library for next-generation WWW”, in Proc. of the 15th Int. Conf. on Web 3D Technology, Los Angeles, CA, Jul. 2010, pp. 165-174.
[9] A. Videla and J. J. W. Williams, RabbitMQ in action: distributed messaging for everyone, Shelter Island, NY: Manning, 2012.
[10] R. T. Fielding, “Architectural Styles and the Design of Network-based Software Architectures”, Ph.D. dissertation, Dept. Comput. Sci., Univ. California Irvine, Irvine, CA, 2000.
[11] P. Hilton, E. Bakker, and F. Canedo, Play for Scala (Early Access Edition), Shelter Island, NY: Manning, 2012.
[12] N. Leroux and S. De Kaper, Play for Java (Early Access Edition), Shelter Island, NY: Manning, 2012.
[13] A. Leff and J. Watson, “Web-application development using the Model/View/Controller design pattern”, in Proc. of the Enterprise Distributed Object Comp. Conf. 2001, Seattle, WA, Sep. 2001, pp. 118-127.
[14] JQuery. http://jquery.com/
[15] S. Weibel, “The Dublin Core: A Simple Content Description Model for Electronic Resources”, in Bulletin of the Amer. Soc. for Inform. Sci. and Technology, vol.24, no.1, pp. 9-11, Oct./Nov. 1997.
[16] M. Doerr, C. E. Ore, and S. Stead, “The CIDOC conceptual reference model: a new standard for knowledge sharing”, in Tutorials, posters, panels and industrial contributions at the 26th int. conf. on Conceptual modeling, Auckland, New Zealand, Nov. 2007, pp. 51-56.
[17] R. Tarjan, “Depth-First Search and Linear Graph Algorithms”, in SIAM J. Comput., vol.1, no.2, pp.146-160, Jun. 1972.
[18] Deep Zoom-Features-Microsoft Silverlight. http://www.microsoft.com/silverlight/deep-zoom/
[19] M. Still, The Definitive Guide to ImageMagick, New York City, NY: Apress, 2005.
[20] Drewnoakes.com – jpeg exif / iptc metadata extraction in java. http://drewnoakes.com/code/exif/
[21] A. Guarnieri, F. Pirotti, and A. Vettore, “Cultural heritage interactive 3D models on the Web: An approach using open source and free software”, in J. of Cultural Heritage, vol. 11, no. 3, pp. 350-353, Jul.-Sept. 2010.
[22] F. Bernardini and H. Rushmeier, “The 3D Model Acquisition Pipeline”, in Comput. Graph. Forum, vol. 21, no. 2, pp. 149-172, Jun. 2002.
[23] Web3D Consortium: Open Standards for Real-Time 3D Communication. http://www.web3d.org/standards/version/V3.3
[24] pdf.js by andreasgal. http://mozilla.github.io/pdf.js/
[25] P. Cignoni et al., “MeshLab: an Open-Source Mesh Processing Tool”, presented at Eurographics Italian Chapter Conf. 2008, Salerno, Italy, Jul. 2008, pp. 129-136.
[26] J. Behr, U. Bockholt, and D. Fellner, “Instantreality — A Framework for Industrial Augmented and Virtual Reality Applications”, in Virtual Reality & Augmented Reality in Industry. Berlin, Germany: Heidelberg, 2011, ch. 5, pp. 91-99.
[27] T. Malzbender, D. Gelb, and H. Wolters, “Polynomial texture maps”, in SIGGRAPH ’01 Proc. of the 28th Annu. Conf. on Comput.. graph.. and interactive techniques, Los Angeles, CA, Aug. 2001, pp. 519-528.
[28]T. Malzbender, D. Gelb, and H. Wolters, “Polynomial Texture Map (.ptm) File Format”, Client and Media Systems Laboratory, HP Laboratories, Palo Alto, CA, Tech. Rep. HPL-2001-104, Apr. 2001.
[29] PTM Web Viewer by Clifford Lyon. http://materialobjects.com/ptm/
[30] Hewlett-Packard labs PTM Desktop viewer. http://www.hpl.hp.com/research/ptm/downloads/download.html
[31] Cultural Heritage Imaging (CHI) RTI Desktop viewer. http://culturalheritageimaging.org/Technologies/RTI/
[32] Inscriptifact Desktop Viewer. http://www.inscriptifact.com/instructions/
[33] J. T. Todd, “The visual perception of 3D shape.”, in Trends in cognitive sci., vol. 8, no. 3, pp. 115-121, Mar 2004.
[34] R. J. Woodham, “Photometric method for determining surface orientation from multiple images.”, in Optical Eng., vol. 19, no. 1, pp. 139-144, Feb 1980.
[35] Imaging Cluster for Archaeology and Cultural Heritage (ICACH), Science and Technology in Archaeology Research Center, The Cyprus Institute, http://imlab.cyi.ac.cy/
[36] IIPImage framework. http://iipimage.sourceforge.net/
[37] ARC 3D Web service. http://homes.esat.kuleuven.be/~visit3d/webservice/v2/index.php
[38] B. Harper. J. G. Hedberg and R. Wright, “Who benefits from virtuality?”, in Comput. & Educ., vol. 34, no. 3/4, pp. 163-176, Apr./May 2000.
[39] L. Marini et al, “Versus: A Framework for General Content-Based Comparisons”, in IEEE eScience, Chicago, IL, 2012.
[40] L. Diesendruck et al, “Digitization and Search: A Non-Traditional Use of HPC”, in IEEE eScience Workshop on Extending High Performance Computing Beyond its Traditional User Communities, Chicago, IL, 2012.
[41] L. Diesendruck et al, “A Framework to Access Handwritten Information within Large Digitized Paper Collections”, in IEEE eScience, Chicago, IL, 2012.
[42] L. Diesendruck et al, “Using Lucene to Index and Search the Digitized 1940 US Census”, in Extreme Science and Engineering Discovery Environment, San Diego, CA, 2013.
About the Authors
Constantinos Sophocleous, Computation-based Science and Technology Research Center, The Cyprus Institute, Nicosia, Cyprus
Luigi Marini, National Center for Supercomputing Applications, University of Illinois at Urbana-Champaign, Urbana, Illinois, USA
Ropertos Georgiou, Science and Technology in Archaeology Research Center, The Cyprus Institute, Nicosia, Cyprus
Mohammed Elfarargy, International School of Information Science, Bibliotheca Alexandrina, Alexandria, Egypt
Kenton McHenry, National Center for Supercomputing Applications, University of Illinois at Urbana-Champaign, Urbana, Illinois, USA


Subscribe to comments: For this article | For all articles
Leave a Reply