by Yoo Young Lee, Erin D. Foster, David E. Polley, and Jere Odell
Introduction
In 2017 the National Library of Medicine (NLM) announced that it would link PubMed records to open access versions of articles in institutional repositories (IRs) through the National Center for Biotechnology Information (NCBI) LinkOut service. This service benefits readers by identifying open access versions of articles that are otherwise hidden behind paywalls. At the same time, LinkOut rewards both the authors that contribute articles to IRs and the IR service itself. Authors benefit from the increased discoverability of their articles and from the increased readership that results when an open access version is easy to find. IRs may also benefit from an increase in web traffic, but also from a general increase of trust and interest in the IR service among potential contributors. In order to participate in the service, IR managers are required to identify articles in their holdings that a) are open access, b) have a citation in PubMed, and c) are not included in PubMed Central (an NLM-managed, public access repository). Mature IRs may include thousands of open access articles. Manually checking each article by searching for it in both PubMed and PubMed Central is time intensive and presents a barrier that may prevent IRs from participating in LinkOut.
In this article, we describe some existing solutions, provided by NLM, that allow for the identification of articles in an IR that are eligible for inclusion in the LinkOut service. We then provide our solution and document the process that we used to export the metadata of over 4,400 open access articles from a DSpace repository and the development of an R script using the ‘rentrez’ package, which provides functions for applying the NCBI Entrez Utilities API (Winter, 2017). Using this approach, we can quickly identify articles that are eligible for inclusion in the LinkOut service, prepare the records, and send them to the NLM. By sharing our process, we hope to provide a useful and accessible solution for IR managers and staff that encourages further participation in, and use of, the LinkOut service.
Background
Indiana University – Purdue University Indianapolis (IUPUI) is a public university campus that offers 243 undergraduate, graduate, and professional degree programs. The IUPUI campus also serves as the main location for Indiana’s dental, health & rehabilitation, nursing, public health and medical schools. The IUPUI campus includes four libraries: Ruth Lilly Law Library, Ruth Lilly Medical Library, School of Dentistry Library, and University Library. All schools on the IUPUI campus use IUPUI ScholarWorks https://scholarworks.iupui.edu/ as an institutional repository. IUPUI ScholarWorks runs on DSpace and is hosted and managed by IUPUI’s University Library. Librarians from all campus libraries participate in providing outreach and assistance with deposit of works in the repository.
IUPUI ScholarWorks provides open access to over 12,000 contributions from IUPUI constituents–including: historical documents, student theses and dissertations, conference proceedings, working papers, reports, and scholarly articles. Launched in 2004, the majority of the initial deposits to IUPUI ScholarWorks were historical documents and student theses and dissertations. In recent years, however, the deposit of scholarly articles by faculty authors has greatly increased. This change is largely a response to IUPUI’s adoption of an open access policy. On October 7, 2014 the IUPUI Faculty Council voted unanimously to adopt an opt out open access policy based on the Harvard Model Open Access Policy https://osc.hul.harvard.edu/modelpolicy/ (IUPUI Open Access Policy, 2014). Under the policy, faculty authors retain their copyrights to scholarly articles and agree to deposit them no later than the date of publication in an open access repository, such as IUPUI ScholarWorks. The policy also gives the library permission to make these deposits on behalf of faculty authors. If authors can not or do not want to participate in the policy, they are given an opportunity to opt out for individual works. In an effort to make participation as easy as possible for faculty authors, articles that are published in open access journals and articles that are uploaded in copyright-permissible forms to open websites are harvested by the library and archived in IUPUI ScholarWorks. PubMed Central is a leading source of accepted manuscripts harvested for deposit in IUPUI ScholarWorks, due to the fact that the majority of IUPUI faculty work in the health sciences. When an open access version of an article cannot be found in PMC, arXiv, or another source, the campus author of the work is notified and asked to deposit or to opt out. As a result, IUPUI ScholarWorks is the only open access source for many of the article manuscripts authored by the faculty. A significant number of these articles are indexed in PubMed.
Relative to the annual output of scholarly articles by IUPUI faculty (~3,000 articles per year), the open access policy workflow and systems are maintained by a small group of people–less than 3 FTE. The library, therefore, has a strong interest in finding ways to reduce the labor of supporting the open access policy while also ensuring that there are few barriers to faculty participation.
What is the LinkOut Service?
NCBI’s LinkOut service allows direct linking between NLM’s NCBI databases (e.g., PubMed, GenBank, dbGAP) to information and services provided by external institutions and/or organizations (LinkOut Help, 2005). This service enables, among other things, the inclusion of links to full-text publications in PubMed citation records. For example, a library can coordinate with NLM to ensure that the library’s subscription holdings are linked to from PubMed, most commonly through an institutionally-specific icon that is displayed in a PubMed citation record. This allows users to access full-text holdings via PubMed based on their institutional affiliation and their library’s subscriptions. In addition to full-text publications, the LinkOut service provides access to other types of information, such as consumer health information, research tools, and links to other biological databases and repositories (LinkOut Help, 2005).
NCBI’s LinkOut service recently expanded to allow linking between PubMed and the publicly available content (i.e., full-text articles) held in institutional repositories (IRs) (Bastian and Kwan, 2016). This allows users to access full-text publications in open-access repositories, provided they are not already freely available from a journal and/or via PMC. For IRs that participate in the LinkOut for IR program, an IR-specific icon is visible in PubMed citation records (see Figure 1).
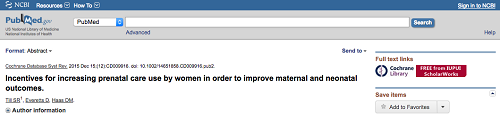
Figure 1. Screenshot of PubMed citation with link to full-text through IUPUI ScholarWorks (see icons)
Requirements for Participating in NCBI LinkOut for IRs
In order to participate in LinkOut for IRs, NLM requires basic information about the repository (e.g., name, URL, contact information), documentation about the IR’s copyright policy, and identification (on the part of the IR) of the number of articles that have a PubMed citation, but do not have an existing publication freely available in PMC. As of earlier this year, IRs that wish to participate in this service must have at least 1,000 articles that fit that criteria (i.e., PubMed citation, not in PMC). This requirement may present the first barrier for an IR’s participation in LinkOut, and readers should take this into consideration before applying to the service or implementing any of the strategies discussed in this paper. The full listing of requirements and contact information for NCBI’s LinkOut is available here: https://www.ncbi.nlm.nih.gov/projects/linkout/doc/IR-application.shtml.
Existing Solutions
There are several methods for identifying the number of items in an IR that have a citation record in PubMed and are not in PMC, but most require a combination of solutions in order to reach the desired result. Additionally, the time intensiveness of certain methods may factor into their feasibility as a solution for some IR managers. In the following, we provide a brief overview of some existing methods in addition to documenting the process involved in developing our solution.
The most straightforward method is manually searching PubMed to determine a) whether an article has a PubMed record and b) if that article is linked to the full-text publication in PMC. Using the PubMed interface is arguably one of the most time intensive methods, particularly since a corpus of over a 1,000 items is now a requirement for participating in LinkOut for IRs.
The following methods are more sophisticated and leverage NLM NCBI tools and APIs to automate portions of the task. This is not a comprehensive listing, but rather an overview of some of the well-documented ways of accomplishing this work:
- Batch citation matcher
This tool uses basic bibliographic information (e.g., author, journal title, year) to find, if available, PMIDs and PMCIDs for citations. This information is formatted into strings that can be entered into a text box on the Batch Citation Matcher https://www.ncbi.nlm.nih.gov/pubmed/batchcitmatch page or uploaded as a separate file. - ID Converter API
This API allows for identification of multiple IDs used by NCBI systems. In this case, using DOIs, one can identify corresponding PMCIDs, if available. The PMCID-PMID-Manuscript ID-DOI Converter https://www.ncbi.nlm.nih.gov/pmc/pmctopmid/#converter tool leverages the ID Converter API https://www.ncbi.nlm.nih.gov/pmc/tools/id-converter-api/ and allows input of IDs (e.g., DOIs) into a text box, which then generates a file of corresponding PMCIDs. If a PMCID is available, the results will also list the corresponding PMID. However, this is limited to articles in PMC so if the DOI is not found in PMC, no IDs are captured. - ESearch query https://www.ncbi.nlm.nih.gov/books/NBK25499/#chapter4.ESearch
Using the ‘ESearch’ utility of the Entrez Utilities API provided by NCBI, one can check if an article is in PubMed and retrieve the PMIDs while also determining whether or not the same article is in PMC. For example,a query can be built using DOIs to search PubMed to identify corresponding articles while simultaneously excluding them if they are already available in PMC (see Figure 2). Results from these search queries are reported as XML.
https://eutils.ncbi.nlm.nih.gov/entrez/eutils/esearch.fcgi?db=pubmed&term=10.1089/neu.2014.3454[doi]+NOT+pubmed+pmc+all+[filter]
Figure 2. Sample ESearch query using a DOI and filter to NOT PMC
While these solutions may ultimately speed up the process of checking IR articles against PubMed/PMC, using any of these methods requires varying amounts of time and effort in learning and establishing a sustainable workflow. In the next section, we provide a detailed breakdown of our solution.
Our Solution
In order to implement our solution, an IR manager must be able to generate a CSV file from the repository that contains the following basic metadata: unique repository ID, a URL to the repository landing page for the article’s full text, and a DOI for the article. Certain repositories may make it easier to export the necessary metadata. For example, DSpace (the repository software used by IUPUI) makes exporting item metadata and embargo dates in the same file difficult. We provide our solution to this problem. [1]
Our solution relies on R, an open source programming language for data analysis. We use two packages that are not part of the base R package, rentrez and plyr, which help us automate the process for looking up DOIs and returning their associated PubMed IDs (PMIDs) and PubMed Central IDs (PMCIDs).
The rentrez package is developed by rOpenSci https://ropensci.org/ and it enables R community users to interact with the NCBI E-utilities https://www.ncbi.nlm.nih.gov/books/NBK25501/ (Entrez Programming Utilities Help) directly in R environment (Winter, 2017). E-Utilities consist of nine server-side programs via web interface including:
- EInfo
- ESearch
- EPost
- ESummary
- EFetch
- ELink
- EGQuery
- ESpell
- ECitMatch
The rentrez package provides functions that communicate with E-Utilities via a REST API. Some of the functions that rentrez package supports include the following:
- entrez_db()
- entrez_search()
- entrez_post()
- entrez_summary()
- entrez_fetch()
- entrez_link()
Among many useful functionalities, entrez_search() and entrez_summary() are incorporated in the R script presented in this article. The first step in our solution is to read a CSV file of articles from the repository that may be eligible for the LinkOut service. Minimal metadata is required for this solution and the CSV file only contains columns for a DSpace record ID, Handle URL, and DOI. When read into R, this CSV file is transformed into a data frame called PMID_PMCID. Data frames are a commonly used data structure in R that allow for the storing of lists of vectors of equal length. Next, two empty columns are added to the data frame. These columns will hold the PubMed IDs and PubMed Central IDs that are retrieved from E-utilities.
PMID_PMCID <- read.csv("FileName.csv", header=TRUE)
PMID_PMCID$pmid <- " "
PMID_PMCID$pmcid <- " "
The next step is to identify PubMed IDs (PMID) for articles available in the PMID_PMCID data frame by searching E-utilities using the articles’ DOIs. The function entrez_search() allows searching a designated database for records containing specific terms. For example, entrez_search(db=”pubmed”, term=”science”) returns 1,957,121 PubMed records that contain a match for the keyword ‘science’.
entrez_search(db="pubmed", term="science") ## Entrez search result with 1957121 hits (object contains 20 IDs and no web_history object) ## Search term (as translated): "science"[MeSH Terms] OR "science"[All Fields]
In our solution, we developed a custom function myPubmedID(). This function searches E-utilities for a DOI and returns the PMID for associated articles that are found. If no articles are found, the function returns a value of ‘None’.
myPubmedID <- function(x) {
Pubmed <- entrez_search(db="pubmed", term=x)
Pubmed_ID <- Pubmed$ids
if (length(Pubmed_ID) == 1 ) {
PMID <- Pubmed_ID
} else {
PMID <- "None"
}
return(PMID)
}
Once the function myPubmedID() is declared in the R workspace, the function is called iteratively using lapply() function and returns either the PMID or ‘None’ for each record in the data frame. The result is saved as a list, which is a specific type of data structure in R. Before the values in this list can be added to the PMID column in the data frame, it must be converted to a vector by using the unlist() function.
PMID_list <- lapply(PMID_PMCID$doi, myPubmedID) PMID_list <- unlist(PMID_list) PMID_PMCID$pmid <- PMID_list
Some of the articles fed into this process are not indexed in Pubmed and ultimately we will ignore these articles. However, for some of the records, when there is no exact match for the DOI provided to the myPubmedID(), E-utilities defaults to searching all fields. E-utilities takes the prefix of the DOI and looks for a ‘best match’, which it usually finds in the PMID field of an unassociated article. For instance, searching for the DOIs 10.1179/2159032X15Z.00000000038 and 10.1179/1059865014Z.00000000074 returns PMID 15017085.
myPubmedID("10.1179/2159032X15Z.00000000038")
## [1] "15017085"
myPubmedID("10.1179/1059865014Z.00000000074")
## [1] "15017085"
This tendency creates duplicate PMIDs for articles that are not actually available in PubMed if their DOI prefix is identical. As an example above, E-utilites takes the DOI prefix for some records and looks for a best match. Our dataset has multiple DOIs with the same prefix and the duplicated PMID is returned for every DOI with the same prefix if this article is not indexed in PubMed. To facilitate their removal, the duplicated PMIDs should be transformed to a value of ‘None’ using duplicated() function from the R base package and count() function from plyr package (Wickham, 2011).
duplicated_IDs <- data.frame(PMID_PMCID$pmid[duplicated(PMID_PMCID$pmid)])
# This returns n-1
colnames(duplicated_IDs) <- c("value")
# Change the column name
duplicated_IDs <- subset(duplicated_IDs, value != 'None')
# Remove None
duplicated_IDs <- data.frame(count(duplicated_IDs))
# Change duplicated IDs to None value
PMID_PMCID <- within(PMID_PMCID, pmid[pmid==23104645] <- c("None"))
In order to get PubMed Central IDs (PMCID), we could apply the entrez_search() that we used in the myPubmedID() function. However, this returns a PMCID only when the item is not under embargo in PMC and an empty value when the item is under embargo. Instead of using entrez_search() for PMCID, we use the function entrez_summary(), which supplies information about an item with the provided parameters database and ID. entrez_summary() provides not only PubMed Central ID of the article but also its embargo date. With the unique PubMed ID identified in the previous step, we can use entrez_summary() in order to get summary information about the article, which includes the PubMed Central ID. For example, a new variable called summ1 contains information for entrez_summary(db=”pubmed”, id=”25622111″) and this returns the following information:
summ1 summ1 ## esummary result with 42 items: ## [1] uid pubdate epubdate ## [4] source authors lastauthor ## [7] title sorttitle volume ## [10] issue pages lang ## [13] nlmuniqueid issn essn ## [16] pubtype recordstatus pubstatus ## [19] articleids history references ## [22] attributes pmcrefcount fulljournalname ## [25] elocationid doctype srccontriblist ## [28] booktitle medium edition ## [31] publisherlocation publishername srcdate ## [34] reportnumber availablefromurl locationlabel ## [37] doccontriblist docdate bookname ## [40] chapter sortpubdate sortfirstauthor
One of the available objects from entrez_summary() is articleids. The variable summ1$articleids shows other article identifiers associated with the PMID, such as DOI, PMCID, RID, EID, and MID.
summ1$articleids ## idtype idtypen value ## 1 pubmed 1 25622111 ## 2 pii 4 2091742 ## 3 doi 3 10.1001/jamainternmed.2014.7667 ## 4 pmc 8 PMC4346513 ## 5 mid 8 NIHMS663485 ## 6 rid 8 25622111 ## 7 eid 8 25622111 ## 8 pmcid 5 pmc-id: PMC4346513;manuscript-id: NIHMS663485;
In order to capture the PMCIDs for articles in the data frame, we create a custom function called myPMCID(). First, this function returns the summaries of all articles by using the entrez_summary() function and the PMIDs identified with the previous function. Then, the myPMCID() function uses a regular expression, with the grep() function from the R base package, to parse the summaries and return the PMCID or a value of ‘None’, if no PMCID is available.
myPMCID <- function(x) {
if (x == "None") {
PMCID <- "None"
} else {
taxize_summ <- entrez_summary(db="pubmed", id=x)
PMC <- data.frame(taxize_summ$articleids)
PMC_ID <- grep("pmc-id:", PMC$value, value=TRUE)
if (identical(PMC_ID, character(0))) {
PMCID <- "None"
} else {
PMCID <- PMC_ID
}
}
return(PMCID)
}
The myPMCID() is declared in the current R workspace before the function is called with lapply() function. This is essentially the same process used with the myPubmedID() function.
PMCID_list <- lapply(PMID_PMCID$pmid, myPMCID) PMCID_list<- unlist(PMCID_list) PMID_PMCID$pmcid <- PMCID_list
In order to meet the requirements NCBI LinkOut for IRs program, the datasets with PMID and PMCID identified are cleaned up. First, the items should be available in PubMed, so the records with a PMID value of ‘None’ are deleted. Second, the items should not be available in PubMed Central, which means the records with PMCIDs should be removed.
# Delete all rows with PMID value None PMID_PMCID <- PMID_PMCID[ which(PMID_PMCID$pmid != "None"), ] # Delete all rows with PMCID value not None PMID_PMCID <- PMID_PMCID[ which(PMID_PMCID$pmcid=="None"), ]
The full code is available in GitHub. https://github.com/yooylee/PMID_PMCID
Conclusion
By using the ‘rentrez’ package for R and the NLM’s National Center for Biotechnology Information (NCBI) Entrez Utilities API, we are able to efficiently and accurately select repository records for inclusion in the NCBI LinkOut for IRs program. After running the script across more than 4,400 repository records, we identified 557 items that were immediately eligible for a linkout from PubMed.[2] We also identified more than 100 items that will be eligible for LinkOut for IRs inclusion when embargoes on fulltext access expire. Processing time for this task in R requires less than 10 minutes. By our estimate, the task of manually searching PubMed using an article’s DOI and recording its eligibility for PubMed LinkOut for all 4,400 records would require more than 22 hours of work. [3]
Librarians without a basic understanding of R and APIs, may face a steep learning curve for using our approach. Learning the basics of R, however, will provide skills that can be applied to other projects. Furthermore, with our R script written and our process documented, we will be able to send future repository works to NCBI with very little effort.
By reducing the labor of participating in the NCBI LinkOut program, we increase the likelihood that authors, readers, and the libraries will benefit from the program. By including a link in PubMed to a free fulltext version of an article in the repository, we are increasing the discoverability of an open access work authored by a faculty member on our campus. At the same time, increased discoverability provides incentives for campus authors to deposit a version of their articles in the institutional repository. In our case, we are already seeing an increase in web traffic to IUPUI ScholarWorks. Soon after successfully sending 557 records to NCBI, PubMed became a leading source of web referrals to the repository–providing 17% of all referrals to the site, second only to the U.S. version of Google Scholar (24% of all referrals).[4] We also expect that repository works included in the LinkOut program will likely see increased downloads.
In sharing our process for identifying eligible repository items for inclusion in the NCBI LinkOut for IRs program, we hope that other libraries, repository managers, and staff will participate in the program. Assuming that an institutional repository meets NCBI’s current requirements for participation, the process that we have documented here can save libraries the tedious effort of manually checking the eligibility of individual items. As more repositories join the NCBI LinkOut service, the increased discoverability provided by the service will reduce barriers to scholarly literature for readers around the world.
Notes
[1] DSpace makes it difficult to export metadata, including DOIs, along with embargo dates for those items. Our approach to dealing with embargoed records is to compare two data frames by the same DSpace ID using the intersect() function. The data frame of the Embargo contains DSpace record ID, Handle and Embargo Date. This allows us to remove embargoed items before processing.
# Load csv file of items under embargo and create a data frame Embargo # Identify items under Embargo and change column name items_under_embargo colnames(items_under_embargo) # Remove items under Embargo from PMID_PMCID data table PMID_PMCID
[2] The 557 items deposited falls below the 1000-item threshold now required by the NLM. When IUPUI began this project, the 1000-item requirement was not in place. We committed to sending additional records on a monthly basis and expect to meet the 1000-item threshold soon.
[3] To estimate how long manually searching PubMed might take, we timed ourselves searching for a random sample of 10 articles from our IR. For each article, we searched PubMed using the article’s DOI followed by the [doi] field tag, ex. 10.5489/cuaj.4090 [doi]. We noted the presence of a PMCID, indicating ineligible articles, and recorded the PMID for eligible articles.
On average, it took us 3 minutes and 19 seconds to search for the 10 articles. Extrapolating from this average, it could take 5 hours 31 minutes and 40 seconds to search PubMed for 1000 articles. This calculation potentially underestimates the true amount of time it would take library staff to complete these searches, as an individual is likely to execute searches at varying speeds throughout a search session. When we compare this with our solution, which processes 1000 records in 2 minutes and 1 second, the time savings are evident.
[4] The LinkOut to IUPUI ScholarWorks in PubMed was officially available on August 8, 2017. According to Google Analytics, 190 sessions came from PubMed between August 8 and August 16 while there was 0 traffic referral in the previous period (July 30 – August 7, 2017). This also increases the overall referral traffic by almost 40%. When we compared the same period of the last year (August 8 – August 16, 2016), there was no traffic referral from PubMed during this time.
References
Bastian H, Kwan Y. 2017. Institutional repository LinkOut: a new full text access feature in PubMed. NLM Tech Bull [Internet]. [cited 2017 Aug 15]; 2017 March-April(415):e3. Available from https://www.nlm.nih.gov/pubs/techbull/ma17/ma17_linkout_institutional_repository_icon.html
IUPUI Open Access Policy [Internet]. 2014. Open Access at IUPUI. Indianapolis (IN): Indiana University – Purdue University Indianapolis; [Cited 2017 Sep 1]. Available from: https://openaccess.iupui.edu/policy
LinkOut help [Internet]. 2005. Bethesda (MD): National Library of Medicine; [Cited 2017 Aug 15]. Available from https://www.ncbi.nlm.nih.gov/books/NBK3805/.
Wickham H. 2011. The split-apply-combine strategy for data analysis. J Statistical Sftw [Internet]. [Cited 2017 Aug 17];40(1):1-29. Available from http://www.jstatsoft.org/v40/i01/.
Winter D. 2017. rentrez: Entrez in R. R package version 1.1.0 [Internet]. Berkley (CA): rOpenSci; [Cited 2017 Aug 15]. Available from https://CRAN.R-project.org/package=rentrez
About the Authors
Yoo Young Lee http://orcid.org/0000-0002-9867-6070 is the Digital User Experience Librarian and IT Analyst Developer at IUPUI. She works with the University Library’s Center for Digital Scholarship to provide skills and knowledge in user experience, application development, project management, and data analysis.
Erin D. Foster http://orcid.org/0000-0001-6908-9849 is the Data Services Librarian for the Indiana University School of Medicine. She works, in collaboration with the University Library’s Center for Digital Scholarship, to develop policies, services, and infrastructure that enable faculty and students to manage, share, and preserve their research data at IUPUI.
David E. Polley http://orcid.org/0000-0003-3595-9708 is the Social Sciences & Digital Publishing Librarian at IUPUI. He works with the University Library’s Center for Digital Scholarship, managing the Center’s open access journal publishing service and providing consultation services in data analysis and visualization.
Jere Odell http://orcid.org/0000-0001-5455-1471 works as a scholarly communications librarian to support open access publishing practices. At IUPUI, he works with the University Library’s Center for Digital Scholarship to implement the IUPUI Open Access Policy, to manage the library’s open access publishing fund, and to provide other library publishing services.


Subscribe to comments: For this article | For all articles
Leave a Reply