By Steven Braun
Introduction
Assessment has become an essential and ubiquitous part of the operations of academic institutions. With increased competition for funding and the unspoken mandate to “publish or perish,” researchers and institutions are increasingly pressured to demonstrate the value and impact of their research scholarship against that of their peers. As the nature of scholarly communication continues to rapidly evolve, demonstrating such impact can come in many different forms, and as a result, both institutions and researchers alike have become burdened with the responsibility of managing all of the varied ways in which their value is represented and compared to that of others.
In this environment, research impact metrics have become particularly popular and widely adopted across institutions. The appeal of these metrics is in their claim to precision and ease of calculation; for example, measures such as the h-index, which describes the minimum number of h citations to each of at least h publications authored by a researcher [1], as well as simpler measures like publication counts and total citation counts attempt to capture a snapshot of researchers’ careers in a singular point value. However, these metrics are built on assumptions that are often overlooked when used as a direct proxy for evidence of scholarly impact, resulting in comparisons between researchers and institutions that may be inaccurate. As a result, properly contextualizing these metrics continues to be a significant challenge to ensuring they are utilized responsibly.
Many platforms currently exist to provide research impact metrics in some form, and each attempts to meet the challenge of contextualization in its own way. Some of these are proprietary extensions of online citation indexes, such as Scopus, Web of Science, and Google Scholar; meanwhile, others are vendor-based or open source and adoptable into existing architectures, such as Elsevier’s SciVal and Pure platforms (formerly called Experts), SciENcv, and VIVO. Each of these attempts to solve the age-old problem of managing the numerous platforms across which researchers’ online scholarly identities may be distributed by providing a standardized and centralized architecture for integrating them all into one location. However, while they are all similar in this functionality, they differ slightly in their approaches, the kinds of scholarship metrics they provide, and the data sources used to calculate those metrics. As a result, it can still be difficult to identify a single platform that may serve all needs around institutional assessment at once.
This problem was encountered at the University of Minnesota in early 2014 when new needs in data-driven assessment arrived at the doorsteps of the University Libraries. Newly appointed in February 2014, the dean of the Medical School was charged with the task of revitalizing faculty scholarship to bolster the Medical School’s international reputation in education and research, attract and retain world-class researchers, and raise the institution’s national rankings. In pursuit of these objectives, the dean sought to programmatically recognize and encourage faculty research excellence through metrics of scholarly output and impact, based particularly on data about their peer-reviewed journal publications. The size and complexity of the Medical School, which employs approximately 2,200 faculty, made this request largely unprecedented in its scope; however, conversations at the highest levels of Medical School and University Libraries administration suggested that the Libraries might have the resources to provide the data needed.
In its earliest iterations, specific Medical School data requests centered around a handful of key data. Delivered in regular reports for each Medical School faculty member, these included
1. A full list of their peer-reviewed publications,
2. their h-index, and
3. metrics calculated about publications on which they were listed as first or last author.
While the first two components were relatively straightforward in their acquisition, the third request for metrics on first or last authorships posed a new challenge. Based on the assumption that having first or last author position indicates greatest contribution to a scholarly work, a newly defined metric named the h(fl)-index sought to capture the frequency and reach of publications substantially authored by faculty. A permutation of the h-index, the h(fl)-index is calculated in precisely the same way as the h-index but with a slightly different set of data: instead of rank-ordering all publications by a researcher, only the subset of first or last author publications by them is ranked by descending citation count and used to identify the highest threshold at which the citation count of a paper is equal to or greater than its rank. This metric, which was defined by the Medical School, is not recorded in existing scholarly literature and as a result required specialized calculation.
As with the h-index, determining the h(fl)-index of a researcher is feasible with complete data about their publication history, from which first or last authorships can be retrieved. Since this prerequisite was already a component of the original data request, responding to the request depended upon a canonical source of publication data for Medical School faculty. For several years, the University Libraries have managed a local instance of Elsevier’s Experts [2] platform (named Experts@Minnesota), which produces profiles for researchers that include lists of publications and other scholarly works, calculations of h-indices, and data about collaboration patterns with other researchers. Publication data are derived principally from Scopus, which is well-representative of the physical, life, and health sciences, disciplines in which Medical School faculty actively publish. Since the majority of Medical School full faculty have profiles in Experts@Minnesota, it seemed that a canonical source of data was found.
However, as specifications around the requested data continued to be refined, further investigation revealed several limitations in using data from Experts@Minnesota alone. The first of these was the discovery of incompleteness in the Medical School faculty list included in Experts. In its entirety, the Medical School faculty population is split roughly in half into two classes. One half includes those with full faculty status, those who functionally practice or work in the Medical School campus proper. The other half, however, includes affiliate faculty, those with primary appointments in the Medical School but who may functionally work at a satellite institution, such as the Veterans Affairs Medical Center. While most full-status faculty were included in Experts, affiliate faculty were largely excluded, meaning that there was no centralized source for retrieving their publication data.
This incompleteness meant that specialized faculty metrics, including the h(fl)-index and counts of first/last author publications, could not be derived exclusively from Experts@Minnesota data. An additional oddity was discovered in the h-index values provided by Experts@Minnesota; rather than being derived directly from h-index values provided on Scopus author profiles, Experts@Minnesota calculations were based only on the limited subset of instances of Experts at other academic institutions, making them unreliable. These limitations had significant implications for how the data the Libraries endeavored to provide would be used: as these data were to be considered in performance review, promotion, and tenure processes, it was imperative that their reliability be prioritized. Additionally, the data reports to be regularly provided were to be used for recognizing top-achieving faculty and tracking faculty publication activity, meaning stakes were high for faculty and the Libraries alike.
In recognition of these limitations, I developed a novel system that could be custom-built to the highly specific needs of the Medical School. Named Manifold, this system dynamically generates data-driven profiles of scholarly output and impact metrics for all Medical School faculty. These profiles include lists of publications, faculty metrics (including custom metrics), as well as visualizations of those metrics, with the aim of providing better context for their interpretation and use. Publication data are principally drawn from Scopus via the Scopus API, and all analytics processes are carried out following a regular schedule to update all data stored internally.
In this paper, I discuss the technical specifications of the system, challenges encountered in its development, and future directions in which development might continue. While the focus here is on the technological dimensions of this project from a systems perspective, a discussion of Manifold from a services perspective will be provided elsewhere (Braun, Manuscript in preparation).
Manifold: Technical Specifications
Overview
Manifold is built on a standard LAMP (Linux/Apache/MySQL/PHP) framework, making it relatively easy to deploy on most architectures. All processes at both the front and back end are written in PHP, and data are stored in a MySQL relational database. This distribution of functionality is based on a model-view-controller (MVC) design that seeks to make Manifold lightweight and extensible. The full source code can be found on GitHub at https://www.github.com/braunsg/manifold-impact-analytics.
The creation of Manifold was spurred principally by two broad needs specific to the Medical School. In one capacity, Manifold serves as the analytics platform needed to calculate research impact metrics and aggregate data about faculty publications for ongoing review purposes. In another capacity, Manifold helps identify potential nominees for the Medical School Wall of Scholarship, a physical installation on the University of Minnesota – Twin Cities campus that recognizes and honors Medical School faculty who have published first or last author papers that have received at least 1,000 citations in two out of three citation indexes (Scopus, Web of Science, and Google Scholar) [3].
As a result, the Manifold system workflow is built primarily with the unique needs of the Medical School in mind. However, Manifold is also built to maximize sustainability, especially in recognition that Manifold data will continue to be used for the indefinite future. The Manifold workflow thus attempts to ensure long-term sustainability by automating all data processes to the fullest extent possible, as described in brief below.
1. Bibliographic data about Medical School faculty, including important parameters such as tenure status, percent time, and job class, are ingested into the Manifold database. On a regular basis, these data are updated for current faculty as well as instantiated for faculty who are newly employed at the University of Minnesota.
2. Using Scopus author identifiers (henceforth “Scopus IDs”), Manifold processes query the Scopus API to pull all publications authored by each faculty member and store them in the database. These publications are subsequently updated at each new data pull to grab new bibliometric data, including citation counts.
3. Research impact and productivity metrics are calculated for each faculty member and stored in the database.
4. Data are output in profiles that are dynamically generated for individual faculty as well as whole departments and custom-defined subsets of faculty.
Figure 1 provides a schematic representation of this workflow. Users only interact with the outputs of the Manifold interface itself, i.e., Manifold profiles.
 Figure 1: Schematic representation of Manifold workflow.
Figure 1: Schematic representation of Manifold workflow.
Profile types and searches
Although Manifold stores data for approximately 2,200 Medical School faculty, profiles for only about half of these — those with full faculty status and who work full time (F.T.E. greater than or equal to 0.67) — are outwardly displayed and thus searchable. (Data for the remaining faculty are stored for other internal review and assessment purposes.) Profiles may be searched for and generated in three views: individual faculty, departments, and subsets of faculty across different departments or job classes that may be custom-defined by users.
Figure 2 provides a screenshot of an example search for an individual faculty member on the Manifold search page. Upon selecting a faculty member to profile, their metrics are previewed for quick reference. Users may subsequently load their full profile for complete data. Meanwhile, Figure 3 shows an example of a custom search, based on selecting a subset of tenured faculty in a given department.
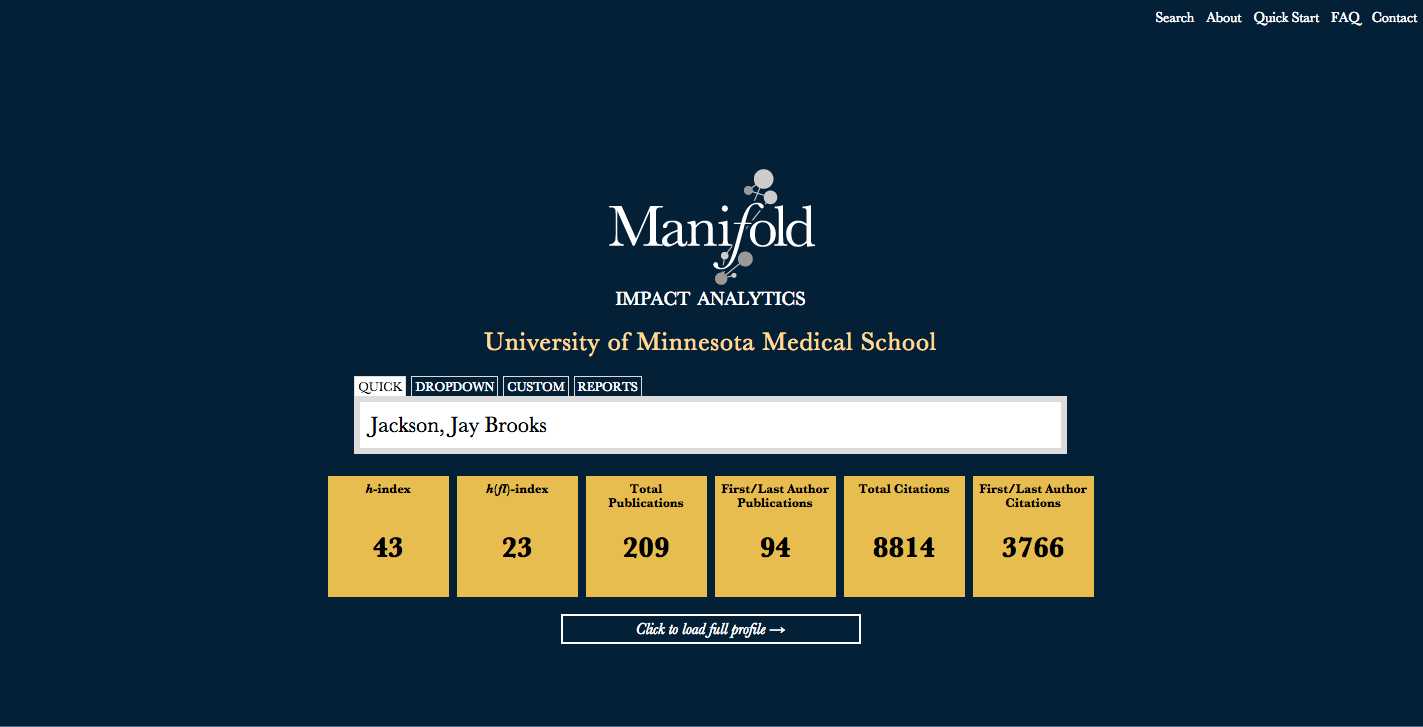 Figure 2: Screenshot of an example search.
Figure 2: Screenshot of an example search.
 Figure 3: Screenshot of an example custom search.
Figure 3: Screenshot of an example custom search.
Profile components
Profiles consist of several components that are dynamically generated based on the entity profiled, each of which is described below.
Metrics
Each profile features an “Overview” module that dynamically displays the following metrics for the profiled entity:
1. h-index, calculated internally based on the publications stored in the Manifold database
2. h(fl)-index, a calculation of the h-index based only on first or last authorships, also calculated internally
3. Total publication count, which may include both Scopus publications (automatically harvested via API processes) as well as records manually imported by users from PubMed (discussed below)
4. First/last author publication count, which may also include both Scopus and PubMed records
5. Total sum of citations to all publications authored by the entity, based exclusively on Scopus citation counts
6. Total sum of citations to all first or last author publications, also based on Scopus citation counts
On individual faculty profiles, these metrics are displayed in conjunction with boxplots that depict the distribution of each respective metric for their home department (minimum, median, maximum, and interquartile ranges) as well as the faculty member’s relative position in that distribution. For departmental and custom profiles, displayed metrics represent median values across all profiled faculty affiliated with the department only, as mean values can be highly skewed by exceptionally prolific and highly cited faculty. Figure 4 provides an example screenshot of these metrics for an individual faculty member.
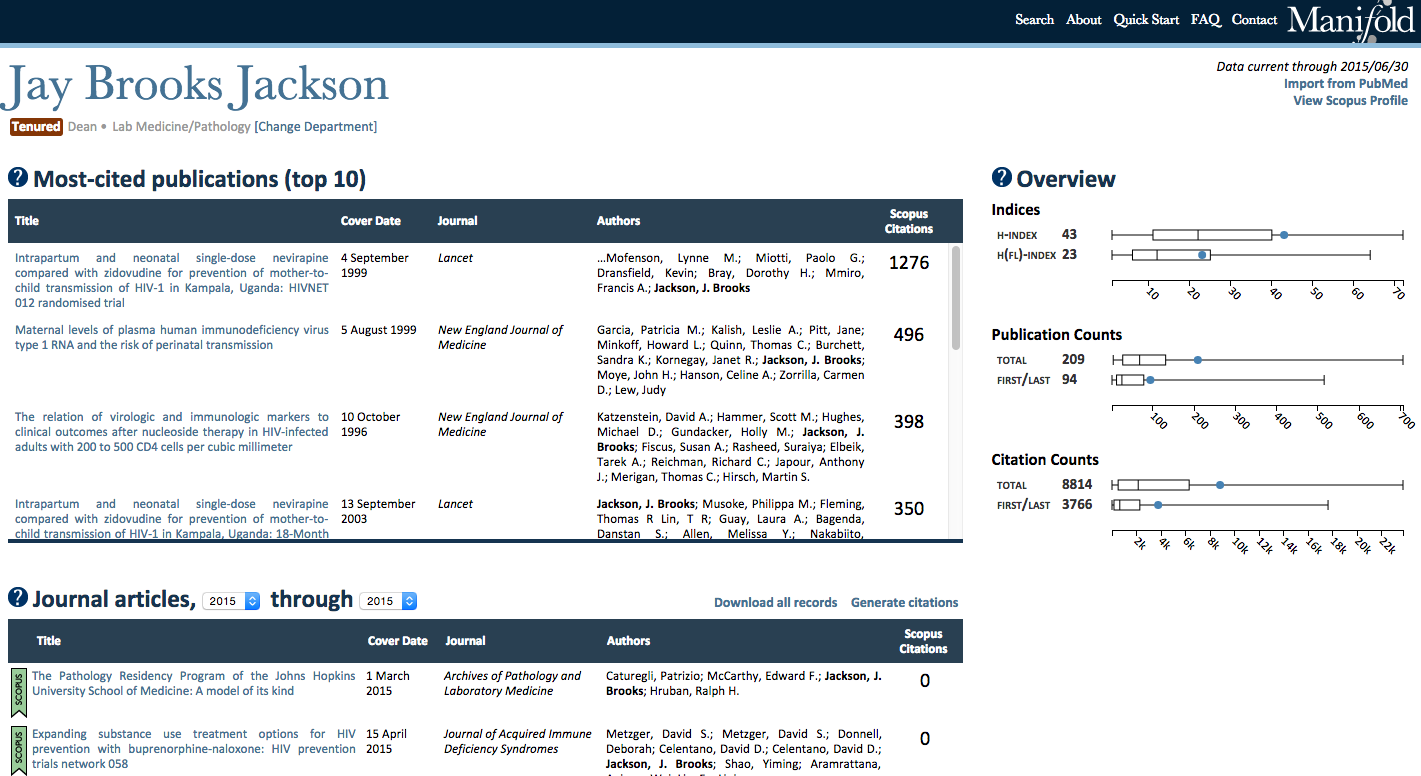 Figure 4: An example screenshot of a faculty member’s metrics.
Figure 4: An example screenshot of a faculty member’s metrics.
Publication lists
Each profile includes lists of publications authored by the entity in two forms, each of which is shown in the example profile illustrated in Figure 4. The first of these lists the top ten most highly cited publications for the entity; the other is a year-delimited list of all publications by the entity, including both Scopus and imported PubMed publications. Users may change the year range of publications displayed, and each Manifold record links back to the original Scopus or PubMed record. For individual faculty, these lists include publications authored by them alone; for departments or custom-defined subsets of faculty, these include publications authored by any faculty affiliated in the defined population. All publication data may be exported to a spreadsheet that can be downloaded by users via the click of a button.
Individual profile publication lists additionally provide users with the option to output the listed publications in a citation format required for University of Minnesota Medical School curriculum vitae. Users may copy this output and paste it directly into other documents. As faculty are being asked to provide metrics and publication data from Manifold in their dossiers for review, Manifold seeks to streamline and simplify this process as much as possible.
Faculty summaries
Department and custom profiles include a table providing summary data for all faculty affiliated with the entity (e.g., all faculty in a single department). This table includes impact and productivity metrics for all faculty and may be exported to a spreadsheet for further analyses.
Visualizations
Manifold is unique from existing systems utilized at the University of Minnesota in its use of visualizations to graphically represent faculty metrics. These visualizations are generated dynamically using D3.js and seek to provide better context for Manifold data that are used for review and assessment purposes. These include distributions of Scopus publications by citation rank order (Figure 5) and year of authorship (Figure 6), with color distinctions for first/last authorships and links back to their respective original records in Scopus; a chart that plots changes in h– and h(fl)-index over time for a faculty member (Figure 7); and comparative distributions of metrics across departments (Figure 8) and the Medical School as a whole (Figure 9).
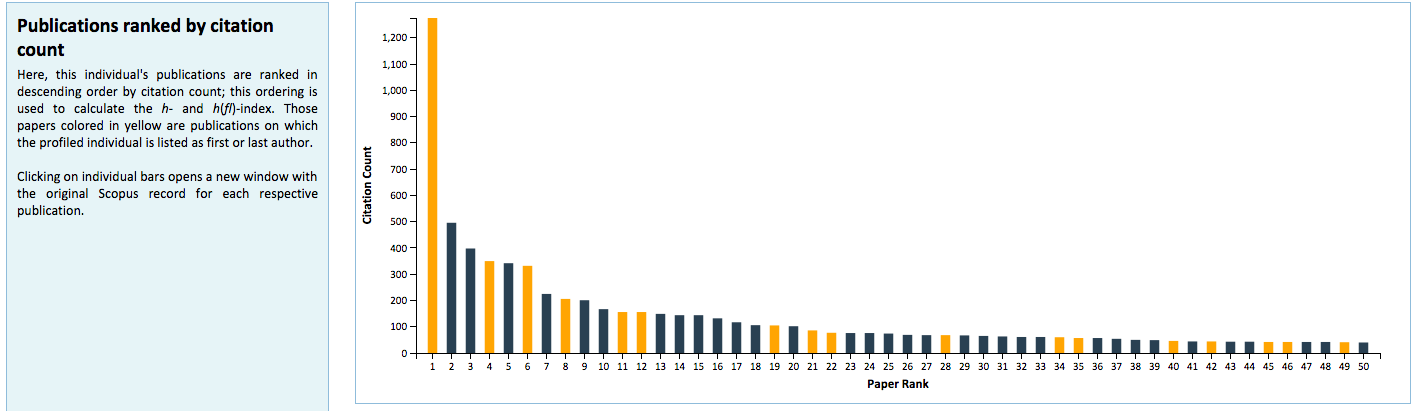 Figure 5: Distribution of Scopus publications by citation rank order.
Figure 5: Distribution of Scopus publications by citation rank order.
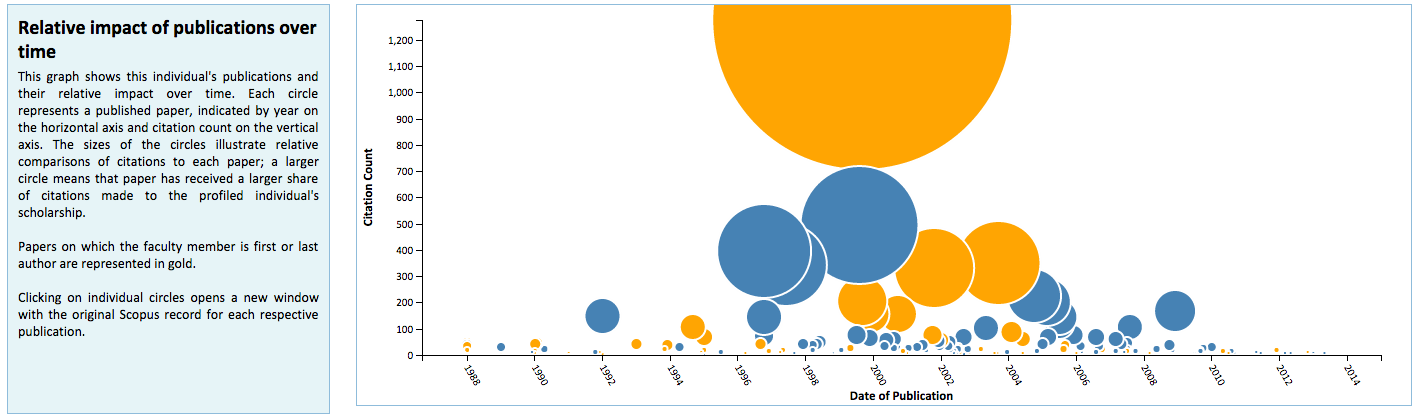 Figure 6: Distribution of first/last authorships.
Figure 6: Distribution of first/last authorships.
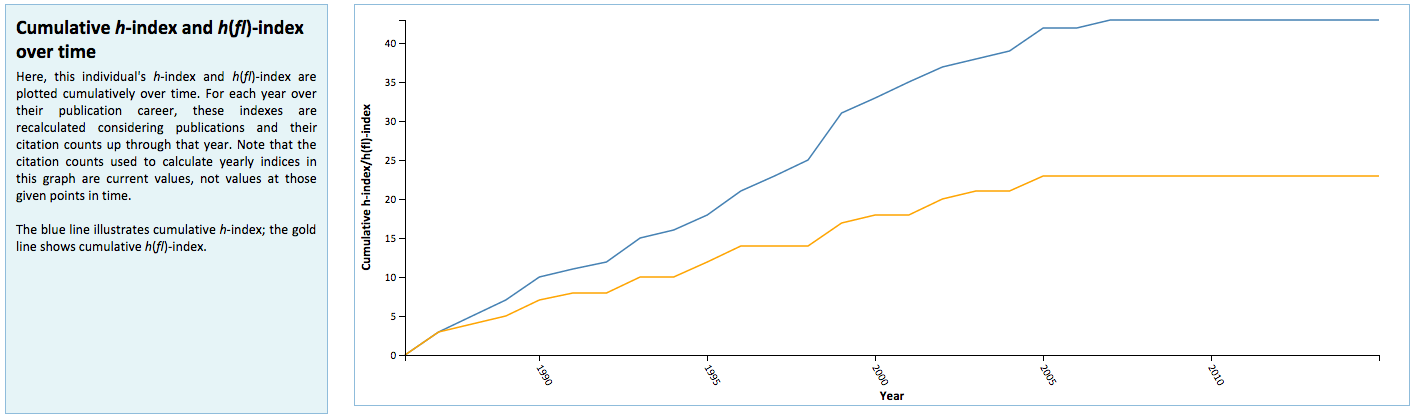 Figure 7: Changes in in h– and h(fl)-index over time for a faculty member.
Figure 7: Changes in in h– and h(fl)-index over time for a faculty member.
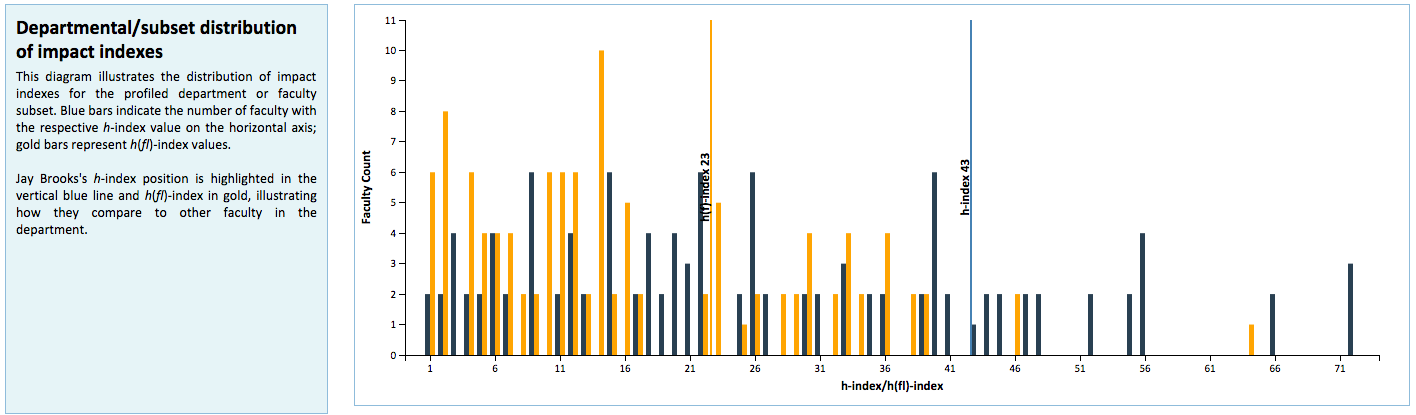 Figure 8: Comparative distributions of metrics across departments.
Figure 8: Comparative distributions of metrics across departments.
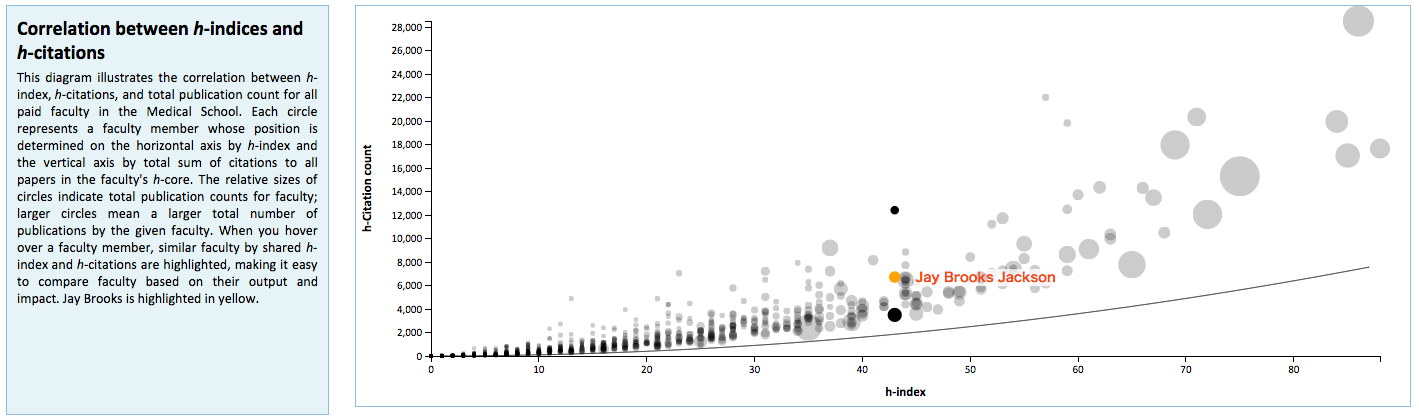 Figure 9: Comparative distributions of metrics across the medical school.
Figure 9: Comparative distributions of metrics across the medical school.
Data
The majority of publication data stored in Manifold are retrieved through the Scopus API, using a registered API key that is credentialed by the University of Minnesota’s licensing with Scopus. Additional data are stored as the result of manual import from PubMed.
Publications
The University of Minnesota Manifold database currently stores publication data for approximately 2,200 faculty in the Medical School. Publications authored by this active faculty pool total over 68,000, which is composed most significantly of publication data harvested directly via the Scopus API. Since Scopus may not index the entirety of a given researcher’s publication history, users also have the option to search for and import missing records into their Manifold profile directly from PubMed, either by title or PubMed ID. This import module is built directly into Manifold profiles, and the process is authenticated via verification codes that are sent to users’ University e-mail accounts.
Scopus author IDs
Faculty publication data are pulled by querying the Scopus API with each faculty member’s Scopus ID; consequently, those IDs must be known in advance. For most faculty in Manifold, these IDs are derived from Experts@Minnesota data, where they exist, using custom scripts that parse the data matched against Medical School faculty. For others without Experts@Minnesota data, Scopus author IDs are harvested via a mixture of manual searching in Scopus, automated scripts that attempt to query the Scopus API based on name and affiliation, and discoveries by human users of Manifold who report incorrect publication attributions or missing publications on their profiles.
Custom reports
Beyond being used to generate faculty profiles, Manifold data have also been used to create custom reports for specific assessment purposes. For example, one such report used at the University of Minnesota is a departmental breakdown of publication counts over time and measures that compare those counts to previous years. Similar reports may be developed for other specific analytics as requested.
Database Structure
All Manifold data are stored in a relational database. Collectively, this database distributes faculty bibliographic data (e.g., name, tenure status, and percent time), faculty metrics, harvested publication records, and links between publications and faculty authorships in records across different tables, each of which holds one of these specific varieties of data.” They also include mappings that transform field codes used in the University central human resources query system, called the Data Warehouse, to internal Manifold field names for interoperability. Descriptions of these tables are provided below.
Table 1: Database structure of Manifold.
| Table name | Table description |
| affiliation_data | Data about (Medical School) departments represented in Manifold, including department codes used in the central University human resources system |
| contact_messages | Data stored about messages sent through the Manifold contact form, for tracking purposes |
| data_sources | Index of data sources represented in Manifold (i.e., Scopus, PubMed) |
| dw_field_mappings | Maps Data Warehouse query field names to corresponding field names in the Manifold database |
| events_master | Data on all processes executed (e.g., publication data update, calculation of metrics) |
| faculty_affiliations | Departmental affiliations of Medical School faculty represented in Manifold, two per faculty: one is the primary departmental affiliation according to human resources, and the other is their functional affiliation in which they are considered active faculty (which may differ from human resources) |
| faculty_data | Bibliographic data on all faculty represented in Manifold, including name, sex, title, tenure status, job code, job class, percent time, and faculty status |
| faculty_identifiers | Various identifiers for faculty (e.g., Scopus ID), one record for each unique ID assigned |
| faculty_metrics | Research metrics for all faculty, one record/row per unique faculty, including all metrics displayed in the Overview module on profiles |
| faculty_publications | Collation of faculty and their authorships; each record links a faculty member with a specific publication and stores data about their position in the author list for first/last author metrics |
| publication_data | All publication data, one record per unique publication, including all bibliometric data retrievable from Scopus or PubMed |
| reports | Data and parameters for loading custom reports displayed in Manifold profiles |
| temp_submissions | Temporary data stored from publications imported from PubMed, before authorized confirmation is made by a faculty user or their proxy |
| visualizations | Data and parameters for loading metrics visualizations displayed in Manifold profiles |
Records in the Manifold database are linked by shared keys between tables. These include identifiers taken directly from the Data Warehouse, such as faculty University internet IDs and department codes, as well as internal proprietary Manifold identifiers. No sensitive or private data about faculty or departments need to be stored as a result of these defined keys, enhancing the openness of the platform overall.
Challenges and Roadblocks in Building Manifold
The development of Manifold in its current version occurred over several iterations spanning approximately 12 months from the summer of 2014 to the summer of 2015. Throughout this course, several persistent challenges emerged at all levels of the Manifold infrastructure, each with its own unique implications for long-term technical sustainability. Here, I discuss a selection of these challenges and approaches to their solution.
Accurately defining a canonical faculty list
Of the many moving parts of Manifold’s architecture, defining a list of faculty to be included in it would seem to be the most straightforward endeavor. However, much to our surprise, this task was the most persistent and resistant roadblock in Manifold’s development. The reasons for this are varied, all related to conventions and idiosyncrasies in the University’s central human resources system that blur the lines between faculty and non-faculty status.
As previously indicated, University of Minnesota Medical School faculty fall into two classes, one for full faculty and the other for affiliates. While full faculty status is defined in a relatively straightforward manner in University human resources, affiliate status is much more complicated, contingent upon many complex variables — including department of primary appointment, line of pay, and location of work. These variables may change rapidly, even over the course of a single pay period, and as a result, a persistent canonical faculty list that is accurate in both its qualifications for inclusion as well as its representation of faculty can be elusive.
An illustration of this difficulty can be found in problems we encountered with the indeterminacy of faculty departmental affiliations. Faculty may have multiple appointments and titles in several departments in the Medical School; some of these may be formalities, related to responsibilities beyond faculty status (such as administrative positions like departmental chairs), and others may be related to the department through which a faculty member’s salary is paid. Due to the transient nature of grant funding, these appointments may flip often, subsequently resulting in significant changes in faculty status. To complicate matters worse, these affiliations also may not always align properly with the department in which a faculty member may functionally work. Since Manifold is designed to run on automated processes, it has no built-in mechanism for detecting these misalignments if appointments are not recorded properly in the original data source (the Data Warehouse).
In early faculty update processes, these inaccuracies were quickly revealed as a result of user feedback. Several faculty reported being displayed in the incorrect department and thus felt inappropriately represented in Manifold. However, these displayed affiliations were derived directly from human resources, meaning that reported inaccuracies needed to be corrected with human resources itself. Given the impracticality of such a solution, Manifold was designed to include a workaround allowing the database to hold two separate affiliation records for each faculty member: one record for their departmental affiliation formally defined by human resources, and one record for their department of functional appointment, to be displayed on Manifold profiles and used for analytics purposes.
Additional problems emerged around even determining which Medical School faculty should and should not be included in Manifold. Early attempts to specify rigid qualifications for faculty status illuminated many nuances in human resources data and sensitivities among faculty; some faculty who were excluded from display in Manifold, perhaps due to their salary appointment, job title, or percent time, felt unjustly misrepresented, even though they objectively failed to meet the basic qualifications we defined. Although making individual exceptions for such faculty was an option, it was not ideal for purposes of sustainability.
To reduce the amount of manual curation needed to clean up faculty lists as a result of these issues, a standard workflow was established that maximizes interoperability with the Data Warehouse while retaining enough flexibility to also define functional departmental affiliations. At each time when the faculty roster requires an update, staff in the Medical School payroll office query the Data Warehouse to produce two lists of faculty, one for full faculty and the other for affiliates. These lists are as rigorously defined as possible, minimizing overlap between them, and include in their consideration corrections for several known Medical School idiosyncrasies that have been revealed due to past experiences. Upon a faculty update, Manifold ingests these lists through a customized series of scripts that update existing or instantiate new faculty records in the database by mapping Data Warehouse query fields to internal Manifold fields. These scripts identify only the relevant fields to import and discard any extraneous data, flagging faculty Manifold records with three fields that declare their status as currently active full faculty or affiliates. After this, additional scripts crosscheck Manifold Scopus author ID records with those provided by Experts@Minnesota data.
Scopus data quality
The quality of any data-driven platform is only as good as the quality of the data upon which it is built. In the case of Manifold, this dependency falls principally upon Scopus, and indeed, the development of Manifold has resulted in a more intimate knowledge of the many different points at which Scopus data quality may fail.
The University of Minnesota instance of Manifold obtains most of its faculty Scopus IDs through data from Experts@Minnesota. However, these author IDs may not always be accurate, resulting in errors that propagate forward into Manifold. Many of these errors are discovered by users of Manifold, several of whom over the past year have reported that their Manifold profile is linked to an author that is not them or lists publications that are misattributed to them. Although Scopus prides itself on its author disambiguation algorithms, in the majority of these cases, these problems were traced back to data provided in Experts@Minnesota, which occasionally assigns incorrect Scopus author IDs to faculty. In other cases, faculty have also reported publications entirely missing from their Manifold profiles. Upon deeper investigation, these absences have been frequently traced back to faculty publications being distributed across several different Scopus author profiles and IDs, requiring a faculty member to merge those profiles so changes in Scopus can propagate forward into Manifold. In yet other situations, Scopus author profiles for faculty may be difficult to locate due to inaccuracies in Scopus author affiliation records. Errors such as these can only be identified and corrected via manual curation, and the Manifold Scopus author ID base has iteratively improved as a result.
Additional oddities have been found at the level of individual publication records in Scopus, particularly in the way that Scopus handles articles in press. When Scopus instantiates a new record for an article in press, that record is assigned a unique Scopus electronic identifier which is subsequently captured and stored in Manifold. Over time, however, those in-press records may be overridden by new records for the full issue-assigned publication, causing the in-press records to disappear. In subsequent data updates, queries against the Scopus API using in-press publication identifiers may thus return errors because the ID has been rendered obsolete and invalid. When this in-press record remains in the Manifold database while a new record for the fully assigned publication is also ingested, the result is a duplication of publication records for individual faculty which consequently affects the accuracy of metrics.
Manifold corrects for this problem by including a boolean flag in the faculty publications table that determines whether a given faculty publication authorship record in Manifold is valid. At the beginning of each full publication update process, all of these flags are reset to zero (false), and as faculty are queried against the Scopus API, only returned records that match against existing records in Manifold are flagged as valid. After a data pull is complete, only valid faculty publication records are used in the calculation of metrics and displayed outwardly in Manifold profiles. Manifold data processes have thus been continuously refined over time to maximize automation and reduce the extent of manual cleanup required afterwards.
Working with the API
Like any API, the Scopus API is proprietarily designed, and a full understanding of its construction, limitations, and fail points can only be developed with experience. Since the foundational functionality of Manifold is derived precisely from interaction with the Scopus API, many challenges in working with it at many different levels have been encountered in the course of Manifold’s development.
One of the earliest challenges resulted from query limits imposed upon Manifold’s registered API key. Although the University Libraries’ institutional contract with Elsevier includes full access to Scopus data for internal use, I encountered persistent blocks upon reaching thresholds around 20,000 query attempts. In the early stages of development, these blocks were a major impediment to the feasibility of the project as a whole; since query limits refreshed on a weekly basis, complete publication data pulls for all faculty — totaling almost 70,000 — would need to be distributed over several weeks. Upon further conversation with representatives from Scopus, these limits were temporarily removed. However, it is unclear whether such limits may be incurred once again in the future.
Additional problems stemmed from arbitrary server errors returned by the API itself. On occasion, queries to the API would arbitrarily fail for specific faculty or publication records due to a general-purpose HTTP 500 error. These errors can only be resolved by halting the process for some time and then attempting the query again, which is impractical when updating a large database of records in sequence. To overcome this, catch mechanisms were implemented to pool failed queries for later retrieval or manual cleanup at the end of entire data processes.
Data load posed several challenges as well. Since the University of Minnesota instance of Manifold currently stores nearly 70,000 publication records by 2,200 faculty, querying each faculty member and publication record in sequence through the API can take significant computing time when run in sequence. For example, in the most recent quarterly update carried out at the University of Minnesota, the process for pulling all new publications for faculty completed in approximately 7 hours; meanwhile, the process to update all existing records completed in 11 hours. These estimates also assume that no errors were encountered along the way. In reality, however, procedural errors can emerge unexpectedly, and thus completing a full data update can span over several days. In the future, these updates may be parallelized across multiple API keys and processes to speed them up.
Prospects for Further Development
In spite of the many challenges encountered along the way, Manifold currently operates smoothly at the University of Minnesota and faculty report fewer and fewer errors in the data it stores. This smoothness is owed to many months of iterative improvements, however, and continued experience working with Manifold will reveal new challenges to be addressed. Manifold is a work in progress, and there are a variety of directions in which it may be developed further to enhance its accuracy and reliability.
Some directions for improvements are currently being explored at the University of Minnesota and may be implemented in new versions in the near future. As the use of Manifold continues to increase in the Medical School, interest in using the underlying data for more custom analytics purposes has continued to grow. One possible direction of growth thus may be the development of an API to allow external access to the underlying data in a controlled, secure connection. Additionally, sources of scholarly works beyond Scopus may be integrated into the Manifold database, including book chapters, educational materials, and other research outputs that may not be captured by Scopus. The resulting heterogeneity would place new constraints on Manifold’s existing database architecture, requiring redesign to handle and represent highly variable modalities of research.
Manifold’s data processes may also be improved in several ways. As has been previously mentioned, publication data pull processes currently query the Scopus API one-by-one, sequentially, resulting in a very long execution time. One possible improvement might be to distribute these processes over multiple simultaneous API connections, allowing for bulk data retrieval. Also, more streamlined mechanisms could be implemented to handle the removal of incorrect data from Manifold. For example, features could be implemented that allow users to manually remove misattributions from their publication lists directly in Manifold, eliminating the need to contact the Manifold developer to remove those data on the back end.
The user interface of Manifold may also be improved in several ways to enhance its user-friendliness. One such improvement may be the inclusion of additional custom user-defined metrics or other ways to visually represent those metrics, and in relation, features that allow users to export or print out those visualizations for other purposes. An additional change may involve providing users with the ability to report co-first authorship on their Manifold publications, a capacity that has been increasingly requested by Medical School faculty in connection to increased emphasis on first/last author metrics. By building in features to enhance flexibility, Manifold can continue to grow and meet new needs as they emerge.
Conclusion
Manifold was custom-built to respond to the specific needs of the University of Minnesota Medical School. Although Manifold has been successful in doing so, the history of its development and challenges therein demonstrates that implementing a system of such magnitude requires careful consideration of the resources available to sustain it for the long term. As other academic institutions explore new platforms to meet their own analytics and assessment needs, it is hoped that Manifold may serve as an example framework that may be adapted, extended, and improved upon.
Code
The code for Manifoldin its entirety is available open source on GitHub at https://github.com/braunsg/manifold-impact-analytics and is protected under a GNU General Public License (v2).
Acknowledgments
I would like to acknowledge Mary Clay, Director of Special Projects (University of Minnesota Medical School), Medical School Dean Brooks Jackson, and Janice Jaguszewski (Director, Health Sciences Libraries, University of Minnesota) for their constant support throughout the course of Manifold’s development. I would also like to acknowledge the efforts of University Libraries Web Development, particularly Michael Berkowski and Cody Hanson, who have put forth tremendous effort to ensure that Manifold can be maintained moving forward. Manifold would not be successful without the support of all of these people in particular.
Notes
[1] For more information about how the h-index is calculated, the reader may find the original citation useful:
Hirsch, J. E. (2005). An index to quantify an individual’s scientific research output. PNAS 102 (46): 16569–16572. doi:10.1073/pnas.0507655102.(COinS)
[2] Elsevier’s Experts platform has recently migrated to their new Pure platform; however, at the University of Minnesota, the name of Experts is still used for reference.
[3] More information about the Wall of Scholarship can be found here: http://www.med.umn.edu/research/wall-scholarship
About the Author
Steven Braun served as the Informatics/Data Services Specialist in the Health Sciences Libraries at the University of Minnesota.


Subscribe to comments: For this article | For all articles
Leave a Reply