By Dave Mayo and Kate Bowers
Introduction
What might seem like a straightforward but sizable data migration can become far less than straightforward when large amounts of legacy data are involved. Both human solutions – like communication and data-driven but fundamentally subjective weighing of possible outcomes – and technical solutions were needed to meet the challenges posed by extreme variations in the data under migration.
In order to understand some of the discussion in this article, a bit of context is necessary. The data we are talking about is metadata in the form of inventories of archival collections. These are also known as “finding aids.” Finding aids are pathfinder documents containing collection-level information and, usually, hierarchically nested descriptions of the subordinate components of an archival collection. Each inventory may have zero to thousands of these nested components, and the nesting is theoretically infinite, although the deepest nesting at Harvard was seven levels. Some elements of finding aids are prose-heavy, such as lengthy family histories, and some are more suitable to being parsed and atomized, such as physical measurements and counts.
In Harvard’s case, the challenge was to move more than 6000 XML-encoded archival finding aids containing two million hierarchically nested descriptions of archival collection components from a native XML database to ArchivesSpace, which is a metadata management system run against MySQL. Harvard University adopted ArchivesSpace (ASpace) in 2015, and decided to move its enormous trove of finding aids from a home-grown backend solution to ASpace. This home-grown backend was called the Online Archival Search Information System (OASIS), and its platform was a Tamino XML server. Originally designed in 1995 (OASIS Steering Committee, 1996), the last significant changes were made in 2002 (OASIS Steering Committee, 2003).
Key to the replacement of OASIS with ASpace was the migration of the descriptive metadata in the finding aids. ASpace has an XML importer for the Encoded Archival Description (EAD) markup that Harvard archivists use when encoding these inventories. This may give the appearance that ingest of the metadata would be fully supported by the new system. However, issues the migration project encountered include scale, variety of the legacy data and the fact that ASpace is far more constrained than EAD, and surprises from the importer.
Scale and Variety of Legacy Data
To understand the nature of Harvard’s legacy data, it will be necessary to place yourself in the shoes of an archivist in the year 1995. Things are looking up. You are feeling grateful to have escaped from an elevator just before Celine Dion started singing for the maybe 700th time that day, and you are excited by the possibilities of your newly-installed browser, Netscape, and the miraculous search engine Alta Vista. You envision replacing a little-used Gopher-browsable list of online finding aids with a full-text-searchable body of these heretofore unyielding texts. As an archivist, you are particularly interested in providing one-stop, full-text searchability to finding aids dispersed in about 40 different archives and libraries all around your campus. Right now, most of these are consulted in hard copy in 40 different locations around campus. The big problem seems to be just getting all that text online and searchable as fast as possible.
You meet with colleagues to focus on a question: do you quickly provide a high volume of text to search, or does that niggling sense that at some point in the future you would regret putting mere html or word-processed documents online convince you to do more? Some of your colleagues advocate the former, since this would be low-cost effort requiring only retyping exactly what is available in hard copy. As a bonus, recently-created inventories are probably still hanging around on diskettes somewhere, so that text is shovel-ready. Others argue that the group should, at a minimum, use marked-up text, like TEI. In fact, an SGML DTD for archival inventories is right now under development. Like many committee decisions, this one chooses a middle ground. The online finding aids will be marked up text, but there will be little central control over how the SGML standard should be implemented by individual libraries and archives. This leaves them free to contribute even if the markup is not ideal. In the end, the Digital Finding Aids Project at Harvard began using the beta version of an SGML DTD called Encoded Archival Description (EAD), with the goal of making contribution as easy as possible by making encoding requirements very minimal.
The minimal requirements for online finding aids at Harvard never changed substantially over many years. These requirements were 1) validity against the EAD DTD, 2) prohibitions against a few formatting elements in EAD such as tables and line breaks, 3) prescribed frontmatter, and 4) presence of a MARC record in the Harvard integrated library system with exactly the same author-title content in the EAD and the MARC. Adherence to additional guidelines was recommended, but optional. (OASIS Steering Committee, 1999 and 2011)
The scale of Harvard’s data determined that any programmatic changes would be done across-the-board. While individual repositories had made their own encoding choices over the years and patterns could be discerned, the pattern of choices were nonetheless far too numerous to implement different programmatic transformations based on patterns that were not discernable across the entire 6000+ corpus. The Harvard University Archives, for example, had nearly 500 finding aids encoded between 1995 and 2016. Staff involved in the encoding ranged from highly experienced archivists to external contractors to interns. Over the years practices and templates had changed. Thus, even within a single repository the interpretation of the standard and encoding decisions were not predictable. Multiplying this situation across all the repositories produces even greater variation. Thus, programmatic transformations that focused on a subset of the corpus of EAD were rejected as impractical, and instead all transformation scenarios had to be applicable to the entire corpus.
Problem Statement and Overall Goals
ArchivesSpace Constraints Compared to EAD
The internal data model of ArchivesSpace varies from and is far more constrained than EAD. These differences presented issues of either “missing” data or data in EAD that had no home in ASpace. To be successful, the migration would need to leave no essential data behind. The first and definitive goal was to make sure that each of the finding aids would ingest, and a close second was to make sure that all the data in the finding aids successfully crossed into the new system. In addition, there was a strong desire to leave no meaning behind, thus a third goal was to ensure that metadata went from its EAD element into the analogous field in ASpace or at least a good second choice.
While data migration was the goal of the project, by no means was it the only goal. Success was also to be measured by the satisfaction of the archivists who made, maintained, and used the data. To this end, archivists were needed to analyze the existing data and determine how completely and how well the migration had met its goals, and to specify desirable outcomes for particular data elements. Technical support for this analysis and specification work was necessary, including help querying the existing XML data and analyzing the resources that would need to be allocated to various programmatic fixes as well as developing an online web “EAD checker” so that archivists could understand how well individual EAD finding aids would fare in the constraints of ASpace.
Because Harvard’s EAD had been growing over twenty years with little prescription or proscription, the scale of the variety was remarkable. Another cause contributing to this variety was that EAD itself made few specific demands. For example, EAD requires that a description contain only one of twelve possible pieces of information: an abstract, or a container, or a link to digital object, or a heading, or the language of the material, or a note, or a creator, or a physical description, or a physical location, or a date, or an identifier, or a title (EAD tag library, 2002). By contrast, ASpace requires a title and date at the highest level in the hierarchy and either a title or a date at all subordinate levels. In addition, there are data elements in EAD that have no home in ASpace. For instance, alternate or parallel titles can be provided in EAD by repeating the unittitle element with varying @type attribute values, but in ArchivesSpace, there is no current provision for alternate titles, and no conversion in place in the EAD importer for such markup. Because of these and other differences, the majority of Harvard’s finding aids would be rejected by the ASpace importer unless they were modified.
In all, archivists analyzed seventy-three issues that resulted in three classes of problems: 1) failure to ingest, 2) ingest with data loss, or 3) failure to place data in the optimal field in ASpace. The project resulted in the following outcomes:
- Fifty issues were resolved by an EAD-to-EAD transformation; additionally the issue was reported to repositories for optional modification of their EAD data by hand
- Eight issues were resolved by reporting them to the repositories for mandatory modification of their EAD data by hand to facilitate ingest to ASpace
- Eight issues were resolved by reporting the issue to repositories for optional modification of their EAD data
- Six issues were resolved by reporting the issue to ASpace which resulted in modifications to the importer
- One issue was an export issue and was referred to the another group for resolution
What determined these outcomes was a combination of the value of the data and our ability to modify it either programmatically or by hand. Data that was a requirement for the EAD importer or that prevented data loss was of the highest priority, while data parsing and transformation that placed it in the optimal field in ASpace was of great but necessarily secondary value. (A full list of issues is in Appendix A.)
Some transformations were relatively simple. Because EAD had both a generic <note> element and an <odd> element (“other”), but ASpace accepts only <odd>, the decision to transform <note> to <odd> prior to ingest was rational. A number of twists were due to the fact that <note> was valid in more places than <odd>, but archivists quickly identified valid locations for <odd> in which the content of former <note> elements would be reasonably placed.
The most difficult transformations were our attempts to take free-form textual content and parse it into firm data values. An example of data that might be deemed “missing” by the ASpace importer is collection-level physical extent. While Harvard’s finding aids might have a very detailed and explicit information, it may not have been tagged in a manner that ASpace expected. In fact, ASpace recognized only about 50% of our expressions of extent, for example:
<physdesc>
<extent>
approximately 25 cubic feet (75 document boxes; 10,000 photographs)
</extent>
</physdesc>
A data transformation that prepended <extent>1 collection</extent> before the <extent> tags for such values was determined to be the best solution. Rejected options included trying to parse these values. Difficulties in parsing included conditions where the numeric value was preceded rather than followed by the measurement (e.g. “Boxes: 6”) or the unreliability of punctuation as a determinant of count and unit of measurement pairs (e.g. “25 cubic feet (75 boxes (25 half-document boxes; 50 flat boxes)). The measurement “1 collection,” while not really useful, is not misleading. Better yet, it is recognizable by archivists as a product of automated processing.
In some cases, transformation was impossible and the only resolution was to send reports to repositories and require that they change their EAD by hand prior to ingest. Such cases included collections lacking titles, for example.
A great effort was made to report out on all transformations so that repositories could take the option of changing their encoding prior to migration. One such case were collection descriptions that lacked dates. Often these were mere failures to provide tagging around dates which had instead been included as part of titles. However, each of these required a fallback transformation scenario. In the case of dates, the group determined that each collection lacking a date would be given inclusive dates from the year 0 to the year 9999.
Container management features[1] were introduced to ASpace towards the end of Harvard’s migration project, so there was little time for a lengthy analysis. In addition, container management is a particularly hands-on and idiosyncratic issue in archives, where storage facilities, storage locations, and shelving varies widely. While we made no programmatic changes to container data, this issue was referred back to repositories to make modifications if they chose, and most indeed elected to do so after hands-on experience with the container management features in an experimental ASpace instance.
In rare cases, there was simply no solution to offer. A number of repositories included notes explaining the meaning of “Controlled access terms,” for example. In this case, there is no home for this content anywhere in ASpace and archivists determined it was not essential, so no action was taken and the data was allowed to be dropped during migration.
Surprises from the Importer
Perhaps the greatest service we did for the community of ASpace users was report a number of issues to the ASpace team. Among these were the failure of the importer to respect the EAD standard’s default behavior regarding finding aid content that had no @audience attribute value. For content marked for neither an “internal” nor an “external” audience, EAD expects that this content is to be shared, but the ASpace importer marked all elements that lacked any @audience value as “internal” (i.e. “not published”). In this case, we were pleased that the project went on long enough for the ArchivesSpace team to provide and to incorporate code contributions that provided alterations to the importer, so that we did not end up with millions of pieces of hidden data that had previously been accessible.
These reports were passed to the ASpace project team via their issue tracking system, which is accessible to member institutions, and via the issue queue attached to the source code, which they maintain in GitHub. Code contributions made by Harvard were submitted by Dave via Github pull requests.
Technical Goals
Provide support to metadata analyst and archival staff in analyzing our Finding Aids
Dave came onto the project after a substantial amount of analysis had been completed, but our corpus was too large, practice too varied, and too few hands comfortable with metadata work were available for it to be exhaustive. Additionally, there’s not a rigorous external definition for what a valid EAD is in context of ArchivesSpace’s importer. EAD is a very permissive standard; many valid EAD structures fail to import, or import with omissions or outright errors. The only machine-actionable criteria for an EAD file’s correctness with respect to ArchivesSpace is the ArchivesSpace importer’s code itself, so there’s not an easy way to measure success in more than binary terms, i.e. “did it import?” A significant part of development time was spent on aggregating and summarizing errors from ArchivesSpace, as well as on writing XQuery and XPath expressions to run over the corpus (pre and post processing). Dave had enough library metadata experience to pursue some of this analysis on his own, and also did substantial work in support of analysis by the Harvard Library’s archivists and metadata librarian.
Improve ingest process
Although ArchivesSpace did have support for batch imports via the interface, it was (and is still at time of writing) flawed in several significant respects, one of which was considered a critical flaw that blocked ingest of the corpus. The critical flaw, which several other institutions have encountered, is that ArchivesSpace batch imports halt on the first error encountered in the batch, rather than skipping to the next file in the batch. While this hasn’t prevented use of the tool for small collections, and the batch importer continues to be a part of our workflow for files too large to import via the ASpace API, it is clearly not usable with a collection of over 6000 files.
Enable reporting on progress
There was a strong desire from senior and local management to be able to measure progress, even if only approximately. Before implementing the tools, the only measure we really had was “does this file ingest successfully without throwing an error, and does it look correct?” Since any single error would cause a finding aid to be rejected, and many errors were present through large parts or the entirety of the corpus, this produced a flat graph indicating stasis, even as many individual errors were found and repaired. To produce something like an honest proxy for our progress, a focus on producing an auditable record of problems found and processing actions taken over time was essential. This had a nice synergy with our support and analysis goals; in practice, support and analysis drove more development decisions, and much of the reporting functionality was deferred to manual generation of reports by the developer and archivists.
Technical Decisions
Error Detection
The first requirement for migration was a reliable method for detecting errors in the EAD files. All of the finding aids started out as valid EAD, so this wasn’t an issue of validation; what was needed was a method to describe and report on particular structural or content patterns within the EAD files.
Schematron is a rules-based XML language for making assertions about patterns in XML documents. Any pattern describable by XPath can be reported on: for example, here’s the pattern we use to detect extents with non-numeric values:
<pattern id="extent-nonnumeric-manual">
<rule context="//*:extent">
<!-- 'extent' elements -->
<assert test="matches(., '^\s*[0123456789.]')" diagnostics="enm-1">
'extent' element content should not start with non-numeric character except '.'.
</assert>
</rule>
</pattern>
It was initially chosen as the mechanism for detecting errors partly on account of the previous work done, and the potential to use it as a “friendlier” error-checking mechanism in the importer. Neither of those reasons ended up being particularly relevant, but Schematron still ended up being a useful tool for checking errors.
Preprocessor
When it came to actually incorporating our work into the migration process, we had roughly three methods available. We could alter code and submit it to ArchivesSpace for inclusion into the core project, we could create a plugin with the changes, or we could transform the EAD files before ingest in ways that avoided causing problems with ASpace’s existing importer.
While we eventually ended up pursuing all three of these methods, we decided early on to focus on preprocessing our EAD files. This decision was based partially on a somewhat negative early evaluation of the ArchivesSpace plugin system and EAD converter, and partially in response to broader decisions related to local processes. The cost of coordination with the ArchivesSpace project was also a concern, and continued to be a concern throughout the project.
There are a number of attributes that led us to initially decide to avoid focusing effort on the ArchivesSpace converter code. On a surface level, the code is, as far as we were able to tell, sparsely documented, and where documented, is documented in terms of its specific handling of an element or elements. The importer is implemented as a fairly minimal domain specific language over Nokogiri::XML::Reader and the custom JSONModel serialization layer defined by ArchivesSpace. Most existing plugins implementing converter customizations involved small rewrites of specific element-handling code, without nearly approaching the scale of the customizations desired by Harvard. In addition, there were concerns related to the lack of isolation in the design of the ArchivesSpace plugin mechanism, particularly as several system upgrades were expected over the course of the work. The final decision was essentially to do as much as possible in the external preprocessor, and to reserve work on the importer for changes that were not resolvable by altering the ingested EAD, or for cases where the converter’s behavior was clearly and unambiguously a risk of data loss or corruption.
An external preprocessor was also considered valuable because it allowed local progress to be decoupled to some degree from the ArchivesSpace system. While “what does it do when ingested” was still the final arbiter of correctness, many issues were able to be diagnosed and analyzed strictly from looking at the finding aids as represented in EAD. The preprocessor initially allowed for turnaround time of roughly five hours to process the whole collection. While this eventually expanded to twelve hours, as more classes of problems were added to the preprocessor, this was still much, much preferable to the three days that a full ingest of the collection into ArchivesSpace required.
As a side-benefit, having the external processor embedded in a web application made it an ideal place to provide self service access to finding aids and reporting. Reporting was repeatedly de-prioritized, and thus was never really implemented, but self-service download of finding aids was very well used and had a positive impact throughout the migration process. These functions are directly responsible for the choice to develop the preprocessor in the Rails web framework.
Tools produced for this project
Schematronium
The first step undertaken was to develop an automated way to check all of the finding aids for errors in a reasonable amount of time (no greater than overnight), with solid reference to point of error (we ended up getting XPath location and line number in file, which was considered optimal). Our initial survey of the Ruby ecosystem revealed a few existing Ruby libraries for running Schematron – unfortunately, most of them were unsuitable for running XSLT 2.0, which (due to regex support in XPath and a few other concerns) was considered preferable to XSLT 1.0. The one library that did use an XSLT 2.0 processor did so by shelling out to Saxon’s command line processor; this was a non-starter because it essentially meant that each file’s processing was preceded by a cold JVM start up, which (in almost all cases) dwarfed the actual execution time of the schematron processing. With over 6000 finding aids, this led to runtimes well outside what was considered reasonable.
At this point, Dave started hunting for a general purpose XSLT-running library, and found Matt Patterson’s saxon-xslt, which directly used Saxon’s Java API, and which was well-documented enough to make contributing fixes and improvements reasonable.
At this point, Dave sank a fair amount of time into working on improvements to saxon-xslt. Specifically, he added support for configuring the underlying parser (needed for line number support) as well as a number of convenience methods related to running XPath. He also wrote a small ruby library, Schematronium, for running Schematron over a file. This library was reused in all succeeding stages of the project, although to date it hasn’t been taken up by ArchivesSpace itself.
EADChecker
At this point, theoretically, work was ready to begin on the preprocessor. At this point, however, Dave proposed and ultimately built a small, single-form web service, the EAD Checker, to allow archivists and members of the working group to run individual EAD files through the schematron on demand. Actually writing the service was less than a day of initial development effort, and a few more days throughout the lifetime of the project. The service consists of:
- A simple form, with a file input for the user’s EAD, and two radio selects for setting various options.
- A help page, whose content is taken automatically from the schematron file.
- On form submission, the user gets back a list of errors found, in XML or CSV formats
The EAD Checker is, in our opinion, one of the better decisions made in the course of our EAD migration. For the cost of three days of effort, we substantially broadened the class of people able to do EAD testing via the current version of our schematron. Additionally, it served as a communication tool. Because it used the same schematron as our other tooling, the EAD Checker help page served as a consistently up-to-date reference to the current state of progress, at least in terms of what problems we could detect and fix in our EADs.
Since the problems in our schematron were primarily general (i.e. corrections required for any ingest into ASpace), we’ve exposed the service to the internet at large, at https://eadchecker.lib.harvard.edu. This required additional work to secure the service against various XML parse vulnerabilities. Many people are unaware, but most XML parsers, if not all, are extremely vulnerable in their default configuration. For more information, please consult OWASP’s XXE page and other security resources.
At least twice as much work went into securing the tool against XML parser vulnerabilities as every other part of its construction combined.
Code for the EAD Checker is located here.
Preprocessor
Despite being primarily a command-line driven XML processor, the preprocessor is implemented as a Rails web application, running on a medium Amazon ECS instance with 30GiB of additional disk space. Both information privacy concerns and the development cost of authentication and authorization led us to make the application available only to Harvard subnets affiliated with the Harvard Library.
The first step to building the preprocessor was to decide how it was going to manage and identify the finding aids it worked over. Our finding aids are uniquely identified by EADID, but it was quickly realized that if we wanted to be able to reliably audit the effects of the preprocessor, we needed to be able to distinguish between different versions of the EAD files, so that we didn’t attribute to the tool changes made by staff to the input files, and vice versa. It was also considered necessary to distinguish between different versions of the schematron file, to prevent attributing changes in error counts to the EAD improving/regressing which were actually due to changes in the definition of errors.
In the preprocessor, these two kinds of files are managed by classes that serve as file registries, SchematronFile and FindingAidFile. On ingesting a file into the preprocessor, the contents are hashed via the SHA1 algorithm, which generates a unique string based on their contents. They are then stored in a directory on the filesystem, named after their SHA1 value with `.xml` appended. Database records (represented in the system by the classes Schematron and FindingAidVersion) are associated with each file, keyed by SHA1, and FindingAidVersions with the same EADID are connected by a database record and class representing all versions of that particular finding aid. This scheme gives a guarantee that any change to a finding aid or schematron will result in a new object, preserving our ability to audit the schematron’s accuracy over multiple versions of finding aids. As a side benefit, this (somewhat) mitigates the disk space costs associated with storing multiple copies of our large XML corpus, as files that remain unchanged between runs are only stored once on the input side.
The schematron file is used directly via XSLT for doing the actual checking, but it is also processed on ingest by the system and each individual assertion is represented as an Issue, identified by a unique identifier pulled from the @diagnostic attribute. Each Issue represents one known problem that exists across the EAD corpus. There are subsidiary database objects for storing concrete information about individual cases of a problem in individual finding aids, represented as a ConcreteIssue class. It was decided, for reasons of reducing programmer effort, not to implement such a registry for output XML. The output from each run is stored in a subdirectory named for the database id of the run.
The application also has a Fixes class, which represents the set of repairs that can be applied to fixes. These fixes are read in from a directory on application start up, and are written in a compact Ruby DSL developed for this application. As an example, here’s one of the smaller fixes:
# Add @type='unspecified' to untyped containers
fix_for 'ca-1', depends_on: ['noempty-1'] do
@xml.xpath("//container[not(@type)]").each do |el|
el['type'] = 'unspecified'
end
end
The DSL is fully documented here. The most complicated part of the fixes is that, because they can be order dependent, a small dependency resolver was necessary to ensure that they ran in the correct order.
The core functionality of the application is located here, and is essentially a single processing pipeline:
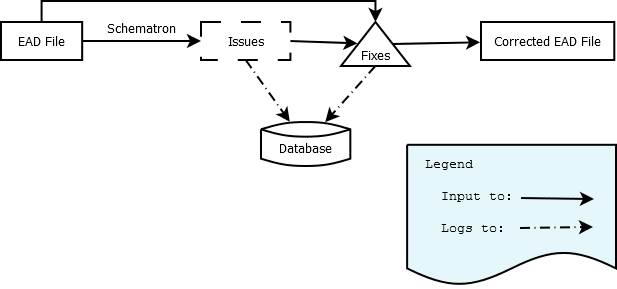
Input EAD is run through the Schematron via Schematronium, with results stored in the DB. These results are used to determine what problems exist in the EADs, and each XML file is then run through the fixes associated with each problem. A record of each fix and its status (succeeded/failed) is stored in the DB. Finally, the altered XML is saved to disk in an output folder.
The lion’s share of the effort put into the preprocessor went into four specific areas:
- The file registries for managing Schematron and EAD files on disk
- The dependency management for ordering fixes
- The methods which actually run the processing over the EAD files
- Automated testing of individual fixes
In order to make the preprocessor generally useful outside of Harvard, an effort has been made to make parts of the preprocessor that are likely to be customized by other institutions pluggable. The schematron, fixes, and tests for fixes are maintained in a separate repository and loaded into the preprocessor on initialization.
The preprocessor is available here, and the schematron and fixes developed and used by Harvard are located here.
Ingest Scripts
Like all other institutions surveyed who migrated from an EAD-based finding aid collection, we wrote an ingest script using the API, rather than use the built-in batch import functionality in ArchivesSpace. The primary driver of this is the batch importer’s behavior around import error. It fails on the first finding aid with errors in it, and, if the error it hits is a Ruby exception rather than a JSONModel validation error, it fails on the first error in the file, which masks all other errors that occur later in processing that file.
There isn’t, as far as we could determine, actually a single “ingest” API endpoint that would handle EAD directly. The endpoint that allows resource creation expects data already in the JSON format understood by ArchivesSpace (and implemented solely in ArchivesSpace). However, Mark Cooper at Lyrasis has written a plugin that accepts an EAD file, runs it through the importer, and returns the resulting JSON representation, or JSON representing the validation or processing error if the conversion failed. So, we developed an ingest script, originally as a set of bash scripts, which were later rewritten in Ruby. The ingest script is located here. It essentially takes each EAD file, posts it to the conversion endpoint defined by the plugin, then posted the resulting JSON to the resource creation API. Errors and their causes were recorded in a set of logs, which were then processed by another script.
While our ingest script does provide the key required benefit (ingest of batch continues despite failure of individual EAD files), the time required for ingest via this method is considerable, averaging between two and three days per complete ingest. Much of the code in our ingest script is the result of an attempt to speed up this process; since a good deal of the time of ingest is spent in network transfer, it seemed like a good candidate for parallel processing, where more than one EAD file could be uploaded at a time. Unfortunately, a race condition in ArchivesSpace (described here) caused failures when more than one resource shared a subsidiary object (agents or subjects, for example) and were uploaded at the same time, and this work ended up producing no real value to the migration project.
Custom Importer Plugin
While we chose to focus on our preprocessor, as stated above, certain issues with import were not solvable by manipulating the imported EAD. In particular, there was no valid EAD structure that would fix ArchivesSpace’s handling of:
- @audience attributes
- <daogrp> elements
- <indexentry> elements
By and large, when writing code for the import plugin, we also packaged it up as a pull request and submitted it to the ArchivesSpace development team; currently, most of the work in our importer is en route to being included in ArchivesSpace 1.5.2.
Credit is due to the Bentley Historical Library at UMich, whose work our <indexentry> fix is based on.
Our import plugin is located here.
Description of our repeated workflow
Our workflow over the course of the past year, once all the pieces in place, has been constructed with repeatability as a primary goal. Working from regular dumps of our EADs as they exist in our production system, we run the preprocessor over the entire corpus. Archivists would then inspect the post-processed EAD to make sure the fixes were transforming the markup as intended. Then, using our utility scripts, we ingested these files into our test ArchivesSpace instance, generally over the weekend, due to the time involved. After the ingest had completed, Dave and the archivists examined the ingest logs and the records produced by ingest. Findings from these sources were used to develop hypotheses as to the cause for classes of failures, and (with cross-checking against the pre-ingest corpus) were used to inform further development.
In summary:
| Action | Work Done By |
|---|---|
| Regular XML dump | automated |
| Run EAD files through preprocessor | developer |
| Analyze preprocessor DB and ouput XML | developer, metadata analyst, archival staff |
| Ingest XML to ArchivesSpace | developer |
| Analyze log from ingest | automated, additional work by developer |
| Analyze corpus (before and after) | developer, metadata analyst, archival staff |
For XML dumps, we made use of standard unix utilities; cron for scheduling, rsync for copying the files to the preprocessor’s host. The full corpus was copied weekly, with incremental updates copied over daily. Throughout most of the work, we worked directly from the weekly dumps, but as we got closer to the final date, we folded in incrementals (manually, using cp over the command line).
The preprocessor and ingest script were run directly via command-line, generally in a screen session to avoid interruptions by terminal timeout or disconnect. This isn’t necessarily recommended practice, or even something that will reliably keep processes alive on some systems; anyone replicating our workflow should make sure to investigate the workable methods of managing long-running scripts on their system.
Individuals involved in the work used a variety of tools and methods for analyzing the XML files; this was less a repeatable, unified process than just people applying what they individually know to the task at hand. Special mention is due to several freely available XML databases, eXist-db in particular. Being able to run XPath and XQuery expressions over the whole corpus was utterly invaluable throughout the work. It was especially useful in finding examples of particular structures, in determining the scope of problems throughout the collection, and in verifying the results of fixes. Additionally, both the official LOC EAD tag library and https://eadiva.com/2/ were invaluable resources for the developer, who hadn’t looked at EAD since grad school.
Retrospective
Good decisions
Schematron ended up being a very good fit for the project. It ended up being flexible enough to describe each of the problems we had.
The EAD Checker proved useful locally, was used outside Harvard, and just generally provided a high reward for relatively low effort and resource cost. It also seems likely to be useful going forward, as an easy way to check ArchivesSpace EAD output, or for institutions who continue to maintain their finding aids in EAD. Additionally, it would be trivial to adapt for other XML metadata formats; all it would take is swapping out the existing schematron for one describing the desired patterns.
Pursuing this process in an iterative fashion by repeatedly running the whole process over the entire corpus on a test system proved to be invaluable. In addition to the intended goals of auditability and reporting, we also got something else useful; a developer with seven months solid practice in cramming EAD into ArchivesSpace. A large number of hard to resolve edge cases came up over the course of our work, that would have been disastrous if we had first encountered them near or during the production load.
While it has the unfortunate result of making the fruits of our effort less accessible to the wider ArchivesSpace community, in retrospect, moving the bulk of our work into a preprocessor and decoupling our progress from that of ArchivesSpace was a good decision. This is not to say that we haven’t benefitted greatly from work done by the ArchivesSpace team and community, but fundamentally, avoiding the need for close coordination with the core team allowed us to keep moving forward during periods of time that the ArchivesSpace team were unable to be fully responsive.
Decisions which in hindsight were not so good
Because the database component of the preprocessor essentially is a glorified log, high quality logging of errors within the preprocessor was left till fairly late in the development timeline. There were several “mysterious” errors which could have been solved much earlier on if internal logging in the preprocessor had been included from the beginning.
While selecting Schematron was overall a positive choice, two decisions made early on in implementation had negative implications for the project. The first, and more minor, was using the XSLT 2.0 version of Schematron, which means it supports XPath 2.0 expressions. XPath 2.0 is significantly more capable, in ways that made describing problems substantially easier at first blush. Key amongst these advantages is its support for regular expressions. But the preprocessor’s actual XML processing code is written using Nokogiri, which is based on libxml2, and only supports XPath 1.0; choosing the XSLT 1.0 version of schematron would have made reuse of expression possible between the schematron code and the processing code.
The second, and more serious bad decision made was in the structure of the schematron document itself. Schematron has two basic reporting constructions, the <assert> and the <report>. Dave made an early decision to use only one of these constructs, to simplify processing of the schematron output. This in itself was probably the right decision; however, he chose the wrong one. Namely, all issues are described via the schematron <assert> construction, which evaluates its XPath in a negated context, i.e. creating output only if the XPath returns false. This, in addition to being arguably more awkward, is the exact opposite of what needs to be done in processing code, where XPath needs to be written to find and return the elements to be acted upon. Almost all bugs written which involved wrong XPath, which made up a substantial percentage of all bugs, resulted from copying and pasting XPath without remembering to negate it, or writing positive expressions rather than negated expressions.
While it didn’t produce problems locally, due to Dave having substantial prior Rails experience, building on Rails probably cost more in complexity for potential adopters outside the university than it provided value in reporting/self-service.
The work spent parallelizing ingest scripts ended up being a complete waste of time, because of the aforementioned race condition in ArchivesSpace. At time of print, the issue is solved in the most recent version of ArchivesSpace, but too late for us to benefit. Every successful ingest completed, including our final production ingest, has been done one file at a time.
About the Authors
Dave Mayo (dave_mayo@harvard.edu) is a Senior Digital Library Software Engineer at Library Technology Services, a division of HUIT at Harvard University, where he has worked for the past three years. He is currently working on behalf of the Office of Scholarly Communications at Harvard, and previously worked as primary developer on the migration efforts described in this article. Dave holds an MLIS from Simmons College.
Kate Bowers (kate_bowers@harvard.edu) is the Collections Services Archivist for Metadata, Systems, and Standards in the Harvard University Archives. She has held a variety of positions in the Harvard University Archives for the past 21 years, all revolving around metadata. Before joining the Harvard University Archives, she worked at Tufts University, the Library of Congress Motion Picture Division, and Harvard Law School. She has also been adjunct faculty at Simmons College since 2009. Her interest is in realizing the potential of metadata and technology to allow people to do what humans are good at–exploring, learning, thinking, teaching, and sharing their ideas.
Notes
[1] Container management is, briefly, a collection of features relating to associating the physical location and containment of materials with their records in ASpace. See http://guides.library.yale.edu/archivesspace/ASpaceContainerManagement and http://campuspress.yale.edu/yalearchivesspace/2014/11/20/managing-content-managing-containers-managing-access/ for more information on the rationale for and development of container management in ArchivesSpace.
References
Encoded Archival Description Tag Library, Version 2002: EAD Elements “<did> Descriptive Identification” (Washington, DC : Library of Congress, 2002) https://www.loc.gov/ead/tglib/elements/did.html accessed 2016-12-01.
Harvard University. OASIS Steering Committee. Digital Finding Aids at Harvard: History and Project Report March 1996 (Cambridge, Massachusetts : Harvard University, 1996) https://web.archive.org/web/20000305224811/http://hul.harvard.edu/hul/dfap/projdescription.html accessed 2016-12-01.
Harvard University. OASIS Steering Committee. Minimal-level EAD v. 1 coding requirements April 1999 (Cambridge, Massachusetts : Harvard University, 1999) https://web.archive.org/web/20000309220757/http://hul.harvard.edu/hul/dfap/dfap_required_tags.html
Harvard University. OASIS Steering Committee. OASIS Annual Report, 2002-2003 (Cambridge, Massachusetts : Harvard University, 2003) hul.harvard.edu/cmtes/ulc/aac/oasis/2003_OASIS_annual_report.pdf accessed 2016-12-01.
Harvard University. OASIS Steering Committee. Harvard EAD Guidelines, revised March 30, 2011. (Cambridge, Massachusetts : Harvard University, 2011) https://web.archive.org/web/20150909021833/http://hul.harvard.edu/ois/systems/mat/eadguide.pdf
Appendix A
Harvard EAD normalization for ingest to ArchivesSpace issues list
| Issue | Description | Resolution | |
|---|---|---|---|
| 1. | field limits | Add check to schematron to report elements that exceed ASpace database field length. | ref to repos |
| 2. | /> | empty elements should be removed in pre-processing so that their presence does not interfere with other tests. | preprocess |
| 3. | <– | Comments are not ingested by ASpace importer. | do nothing |
| 4. | @audience | Audience (internal vs. external) import does not adhere to EAD standard of no value = external | fix importer |
| 5. | arrangement | <arrangement> embedded in <scopecontent> exports invalid EAD on roundtrip. <arrangement> imported twice: once as mixed content within scopecontent and again as a Note with Type=Arrangement. The Note is mangled, only including items from the last, lowest level of a nested list. Preprocess to extract <arrangement> from <scopecontent> and place after. | preprocess |
| 6. | bioghist | On import to/export from ASpace, if there is a <persname> within (or adjacent?) to a <bioghist><p>, all the <p> tags are removed upon ingest. It is also making it invalid on EAD export from ASpace, since <bioghist> needs subordinate elements to be valid. Even if it were valid, it would be one inscrutable block of text. | preprocess |
| 7. | bioghist | If there is a <p audience=”internal”> there is no closing </p> tag, thus the EAD is invalid upon export. | preprocess |
| 8. | c | <c> with no @level import with @level=otherlevel but no corresponding @otherlevel value. Supply @level=otherlevel @otherlevel=”unspecified” in pre-processing. | preprocess |
| 9. | change | If there are multiple changes in our finding aid, multiple revision statements should be created upon ingest. Currently ASpace seems to create a single change statement for all changes. | importer fixed |
| 10. | change | During import, add a <revisiondesc><change> to indicate preprocessor altered finding aid. | preprocess |
| 11. | chronlist | <chronlist> embedded in <bioghist> and <scopecontent> export twice, once embedded in note and second as standalone element. The standalone element should be omitted. | preprocess |
| 12. | container | Analyze uses of container; parse text (e.g., “box”) from data (number) where applicable. WG decided not to normalize this data. |
ref to repositories |
| 13. | container | ASpace containers require a type attribute, but less than 6% of Harvard containers include a type now. Absence of this attribute blocks ASpace importer. Add “unspecified” to controlled list in ASpace and add the value during pre-processing. | preprocess |
| 14. | container | Set @label=”unspecified” if @label is not already present. | preprocess |
| 15. | container | Relationship between container and unitid? Moot unless we parse containers, which we aren’t. | do nothing |
| 16. | controlaccess | Investigate omitting archdesc/controlaccess during migration and instead pull 6XX and/or 7XX from MARC records. | do nothing |
| 17. | dao | Harvard <dao> elements lack xlink:title attribute required by ASpace. For each <dao>, look for one of the following elements in the immediate parent <did>, in this order: <unittitle>, <unitid>, <container>, or <unitdate>. Repeat the text in that element as the <dao> xlink:title attribute. | preprocess |
| 18. | dao | Random ID is being created when <dao> is imported; may need to import existing ID. | preprocess |
| 19. | dao | multiple daos in a <did> or <c> load as separate D.O.s, export as separate daos in the did, but lose their position relative to other elements in the <did>. They are also indistinguishable on export from daos derived from daogrp/daolocs. | ref to repository |
| 20. | daodesc | The <daodesc> is put into a general note AND is not exported in the EAD. | preprocess |
| 21. | daogrp | ASpace imports portions of the component <daoloc>s from an incoming <daogrp> as related File Versions. However, it omits xlink Actuate attribute “onload” and xlink Show attribute “embed” from the associated <arc> element. Preprocess <daogrp> to convert <daoloc>s to <dao>s, take xlink:actuate and xlink:show attributes from related <arc> and add them to the appropriate <dao>; remove <daogrp> wrapper. When exported, two <dao>s in a <did> should be converted back to <daoloc>s with <arc>s in a <daogrp>. | preprocess |
| 22. | descgrp | Descgrp is not supported in ArchivesSpace, and needs special handling to insure that its constituent elements are imported. | preprocess |
| 23. | did | Absence of unitid at the collection level causes import to fail. Supply <unitid> content from MARC record ID. | preprocess |
| 24. | did | Absence of unittitle at the collection level causes import to fail. | ref to repository |
| 25. | did | Absence of unitdate at the collection level causes import to fail. Supply based on earliest and latest NNNN values in component-level unitdates. If no unitdates, supply 0000-9999. | preprocess |
| 26. | did | Absence of both unittitle and unitdate at a subordinate level causes import to fail. Provide unittitle or unitdate from parent component’s <did>. | preprocess |
| 27. | did | Where <did> lacks both unittitle and unitdate, but includes <note>, further analysis needed to identify pattern for conversion. | preprocess |
| 28. | dimensions | dimension element is loaded twice, once as a note w/type=Physical Description and again as a Note with Type=Dimensions. Should be loaded in Dimensions in ASpace Extent when parent <physdesc> includes a simple <extent>. See: https://archivesspace.atlassian.net/browse/AR-1134 | fix importer |
| 29. | extent | Extent is required at the collection level, but is missing in some finding aids. Supply extent of “1 collection.” | preprocess |
| 30. | extent | Separate multiple extents are sometimes encoded in a single extent element. Demote these to <phydesc> which will ingest as physical description type notes by the ASpace importer. | preprocess |
| 31. | extent | Extent needs to be parsed to correctly import whole vs partial extent expressions. WG decided not to parse extents to this degree. | do nothing |
| 32. | extent | Assess each <extent> field for parsability. If parsable, determine conformance canonical pattern and measurement term. Convert alternate terms to canonical term. | preprocess |
| 33. | extent | Multiple extent elements in a single physdesc are loaded with one as a single extent with the others in a container summary. Where extents are truly parallel, can/should they be loaded as separate extents in ASpace? | do nothing |
| 34. | extent | Extents that begin with a statement of approximation (i.e., approximately, circa, ca or ca.) will not load as <extent>. Extract statement of approximation. Process remainder of extent according to rules for extent processing (see above). Add new, additional <physdesc>Extent is approximate.</physdesc> | preprocess |
| 35. | extref | Extref (links) may not import, but the text of the extref will import. | do nothing |
| 36. | index | In some finding aids, indexes are nested, which is not supported in ASpace. In some indexes, the key for implied data within the index entries is provided, e.g., “Key: No symbol = Writer * = Writer and recipient = Recipient” If these indexes are retained, this may need to be used in conjunction with the index entries to make the data explicit. | preprocess |
| 37. | indexentry | An index entry with a ref@target and ref@text imports as two separate items. It should import as one item with associated reference and reference text. | preprocess |
| 38. | indexentry | Nested <indexentry> elements import in order, but the nesting is lost. Repositories have been notified about what will happen if they do not modify EAD. | ref to repository |
| 39. | language | The importer appears to be including Use-For forms of the language in addition to the preferred form. | preprocess |
| 40. | language | Language is not repeatable in ASpace and should be. This may require changes to ArchivesSpace for resources and archival objects. | do nothing |
| 41. | language | Multiple languages in a single language element need to be parsed into separate element occurrences. | preprocess |
| 42. | list | Elements with lists are mangled during import. | preprocess |
| 43. | list | Nested lists (i.e., list/item/list) drop data during import. Solution is to flatten them. | preprocess |
| 44. | list | <list> inside <bibliography> is imported as separate <odd> element, but with titles stripped out. Bibliography note created in ASpace, but doesn’t contain any data. To put it another way: <bibliography> with <head> and a <list> of <items> becomes two notes (General with list items; and Bibliography with head). Drop the <list> element and convert its <item>s to <bibref> | preprocess |
| 45. | name | The importer does not match the documentation: testing for nesting with origination and controlaccess is not occurring, and creation of name_person and agent.type and agent.role is not occurring. Retest after QA version is updated. | report error |
| 46. | name | @encodinganalog is not used to set indexentry (aka Item) Type. Convert <name> to different element based on encodinganalog values. | preprocess |
| 47. | namegrp | ASpace data structure does not support <namegrp> (a structured string akin to a precomposed subject heading). The elements within <namegrp> import as independent index entries, which is inaccurate. | preprocess |
| 48. | note | 1. Note elements are stripped from components during ingest by the ASpace importer 2. ArchivesSpace does not support the <note> tag; importer strips them or converts them to <odd> depending on the context. | preprocess |
| 49. | note | <notes> within <controlaccess> are dropped during import. | do nothing |
| 50. | note | <notes> within <archdesc> are dropped during import. Convert <note> to <odd> prior to load. | preprocess |
| 51. | note | convert <note> to <odd> when <note> is a child of <c> | preprocess |
| 52. | note | when <note> appears as child of <did> convert to <odd> and place outside and immediately following the <did> in which it occurred. | preprocess |
| 53. | note | <note><p> occurs instead of proper element, e.g. <scopecontent> or <bioghist>. | ref to repository |
| 54. | p | Initially there was import/export mangling of <p> resulting in invalid EAD on export. importer fixed, but sibling elements that are not paragraphs may be invalid on export. | ref to EAD export group |
| 55. | persname | @role=”Donor (dnr)” is imported as Role=Subject. | preprocess |
| 56. | physfacet | Same as dimensions. | fix importer |
| 57. | ptrgrp | ref value + target structure is not repeatable in indexentry in ASpace. | preprocess |
| 58. | ref | <unitid> loses @id attribute on load. This makes ref targets that reference unitid @ids invalid. | preprocess |
| 59. | relatedmaterial | See <list> and <extref>, above. extref works, lists depends on resolution of list issue. | preprocess |
| 60. | @startYear/endYear | startYear and endYear attributes in <date> and <unitdate> elements are Harvard only, invalid in canonical EAD used by ASpace. Transform to @normal. | preprocess |
| 61. | @startYear/endYear | When date values represented in startYear/endYear (or normal) attributes are irrational (the start is later than the end), delete the attributes. | preprocess |
| 62. | table | Only 3 Harvard finding aids contain tables. They are not supported by ASpace and should be manually removed prior to conversion. | ref to repository |
| 63. | title | <title> within <controlaccess> is dropped during import. Should be manually changed to subject, corpname, or other choice. | ref to repository |
| 64. | unitdate | Parse descriptions in and around <unitdate> to correctly tag bulk and inclusive dates. | preprocess |
| 65. | unitdate | Sometimes the words “bulk” or “inclusive” inside <unittitle> but not inside <unitdate>. | preprocess |
| 66. | unitdate | When <unitdate>contains both “bulk” and “inclusive”, it needs to be parsed into two separate unitdates in ASpace. | preprocess |
| 67. | unitdate | unitdate includes two date ranges, first with no designation, second with “bulk” before or after date range | preprocess |
| 68. | unitdate | If <unitdate> contains “circa”, “ca.”, “ca”, “approximately”, “approx.” (all case insensitive), set @certainty=”approximate” | preprocess |
| 69. | unitdate | <unitdate> lacks @startYear, @endYear AND @normal. With no action, these will be loaded as Date Expression, with no normalized, searchable dates. Where <unitdate> lacks all these attributes, would it be desirable to examine each NNNN inside <unitdate>, keeping the earliest and the latest and creating a @normal=”[earliest/latest]” | preprocess |
| 70. | unitdate | Because ASpace has separate fields for unittitle and unitdate, it takes rest-of-date information that follows the EAD <unitdate> and includes it in the unittitle, i.e., puts it BEFORE <unitdate>, in non-DACS fashion, with an added comma. | preprocess |
| 71. | unitid | Importer converts “.” to “_” in <unitid> | importer fixed |
| 72. | unitid | For multiple unitids, ArchivesSpace retains the last one and discards the rest. Concatenate with a double dagger separator. | preprocess |
| 73. | unittitle | Multiple <unittitle>s do not import. E.g., one unittitle in English, one in Chinese–only the latter/last unittitle imports. Concatenate with double dagger separator. | preprocess |


Subscribe to comments: For this article | For all articles
Leave a Reply