by William Roy and Chris Gray
Introduction
The Confederation of Open Access Repositories (2012) acknowledges that, “most content recruitment practices are fairly resource intensive and involve staff resources” (p. 2). At the University of Waterloo, mediated deposit is a multi-stage process consisting of communications and promotions to researchers, review of faculty CV’s for eligible works to deposit, checking for copyright and licensing considerations, and the manual entry of metadata in order to be consistent with local repository metadata standards. Mediated deposits have proven to be a useful way to stimulate usage by faculty. However, this option requires a far greater investment of time for library staff than self-depositing workflows require, because users are not required to enter metadata at the time of ingest. At the University of Waterloo, it was discovered that staff were spending close to 50% of their time on the manual collection, clean-up, and entry of publisher metadata into spreadsheets. This part of their mediated deposit service required rethinking so that staff could spend more time on the rest of the outreach and copyright-related activities required for recruiting content and less time on data-entry.
There have been numerous and notable efforts made by institutional repository (IR) managers and technologists to streamline the collection and harvesting of publications’ metadata. IR’s collect and provide access to a large variety of original scholarly works, but a primary focus still remains on providing access to published works with ‘green’ open-access friendly versions, or openly-licensed publisher versions that are free to redistribute and share on their platforms. The accompanying descriptive metadata needed for most repository objects is already existent in some shape or form in citation or registration agency databases, but is likely in need of transformation for ingest into a repository. Examining the previous literature demonstrates the varying lengths to which repository personnel have gone to retrieve and transform data into formats useful for their local context. The College of Wooster used an XML export of selected citations from the commercial program RefWorks and transformed the resulting citations metadata into a repository-ready Dublin-core XML format using custom-written XSLT (Flynn, Oyler & Miles, 2013). Yuan Li (2016) successfully used SoapUI with the Web of Science web services to harvest publications metadata in XML and similarly employed a custom XSLT to transform the output so that it conformed to local metadata standards. Finally, a most recent and robust solution from Powell, Klein and Van de Sompel (2017) automated metadata collection, along with several other aspects of the content recruitment process by using ResourceSync, Django Solr sync, multiple API’s (e.g. Sherpa Romeo, Crossref & OADOI), and a Neo4J graphing database.
Rationale
We considered similar routes as discussed above, but due to the nature of how different citation services package and output metadata, it was found that no single source (e.g. Web of Science, Scopus, and various citation management software packages) could be relied on to consistently contain the fields that were of interest to our local repository schema. Of considerable challenge were metadata containing funding references, Creative Commons licenses, and affiliation information. XSLT, although a powerful timesaver, is not easily editable for those not trained in the language. We assumed using XSLT would sacrifice support staff’s ability to manually edit or supplement retrieved metadata that was not ideal upon initial collection. Support staff who edit repository metadata may not have the training to dissect potential problems and edit the resulting XML documents but would be much more proficient working with spreadsheets.
The method of using spreadsheets to organize metadata for import into various repositories and information systems can be a popular choice for libraries. Due to the low barrier for adoption by new and contract staff members, using a data-entry workflow that consists of entering metadata into spreadsheets was a natural choice for our library. DSpace, the repository used at the University of Waterloo, utilizes a tool called the SAFBuilder [1]. SAFbuilder, available in Bash script or Windows batch file format, accepts a CSV document’s filename as its argument and packages metadata and file names indicated within the spreadsheet into separate DSpace ready archive folders, complete with properly-formed metadata in XML. Many other repositories have similar options for batch metadata entry. For example, Digital Commons offers a “Batch Upload Excel” option [2], and Islandora modules exist for importing MODS metadata through CSV [3].
It is our hope that by utilizing a method of semi-automated metadata collection in harmony with a typical spreadsheet workflow, we can provide a relatively platform-agnostic approach that is simple for support staff to use and to alter as needed. In addition, because this method allows for communication with APIs, it remains flexible enough to connect to other emerging scholarly services. There are a variety of interesting services arising and growing each day that source value-added scholarly metadata using APIs such as: ORCID, the Scholix: Framework for Scholarly link Exchange, Sherpa Romeo, and Datacite. Since the urllib Python package that the script uses is able to make web requests and the json package can communicate and parse JSON data, this method, given some additional tweaking, could also allow for easy association of items to things such as: linked datasets, ORCIDs, and Sherpa Romeo information via the list of DOIs supplied to the script.
Defining the Problem
As mentioned earlier, no single source was all-encompassing in offering the type of metadata fields our repository hoped to populate. Some pain-points in particular were:
- Funding Information
- Creative Commons licenses
- Affiliation Information
- Full bibliographic citations
Citation software, such as Zotero or RefWorks, do not capture these fields by default. We found Web of Science (WOS) to be the most complete source of information that staff would rely on for the retrieval of metadata [4]. Despite being able to export a TSV from WOS with a reasonable amount of the metadata we required, the above fields were either non-existent in WOS, or required further editing to be ready for packaging by SAFBuilder. Additionally, the TSV exported from WOS contained numerous extraneous columns that were of no use.
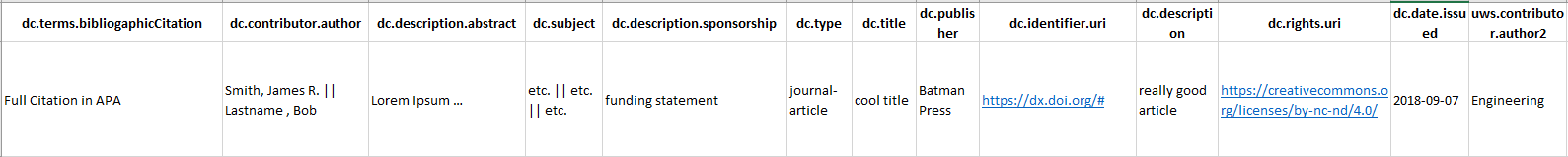
In summary, we wanted to attempt to populate all relevant fields (Fig. 1) with metadata that we deemed consistent with our local standards and specifications.
Figure 1. Column structure showing metadata fields our script targeted. Click to enlarge
Before implementing our Python script, the workflow [5] was as follows:
-
- Locate a list of titles on WOS that are ready for import into the repository and add them to a marked list.
- Use the marked list parameters to select all applicable metadata fields to capture.
- Export a TSV file of the marked list.
- Open the TSV as a spreadsheet and copy and paste the fields of interest into a SAFBuilder template, avoiding extraneous columns. The column headers of the SAFBuilder CSV represent Dublin Core field names, so staff must discern the appropriate matching column in the WOS TSV, and copy and paste accordingly.
- After transferring useful information from WOS TSV, identify missing metadata and fill in remaining fields manually by locating the information on publisher’s website.
- Manually change formatting to be compliant with local standards (i.e. switch semicolons to double pipe symbol, use concatenation formula to make DOIs resolvable, alter date fields to display in one column rather than two, and use ISO 8601 format).
- When complete, run the SAFBuilder script with the newly created CSV and import the resulting ZIP folder to UWSpace.
Our Proposed Solution
Overview of Method
The scripts use standard Python libraries with the exception of the third-party Habanero [6] library. This library has a Crossref module that is imported to request JSON data from the Crossref API. PIP was used to install Habanero.
Alternatively, if you do not want to install Habanero, it is possible to access the Crossref API by just using the standard urllib.request library.
The Python script starts from a spreadsheet (tab delimited, utf-8) downloaded from WOS. The WOS metadata export should include the following fields. We prefer to obtain author information, keywords and abstract metadata from WOS because information for these fields is often more complete, or formatted in ways that is preferred by our repository. DOIs are obtained at this step for the purposes of creating the Crossref lookup, and titles are obtained as a useful point of reference for troubleshooting if the DOI is found to be non-existent. The WOS metadata export should include the following fields:
- Author
- Title
- Author keywords
- Keywords Plus ®
- Abstract
- Digital Object Identifier
These fields appear in the tab delimited file as: AF, TI, DE, ID, AB & DI.
The script then takes each DOI from that data, and uses the DOIs to get citation information for each record using Crossref content negotiation [7]. It – obtains supplementary metadata fields for each record from Crossref by converting the JSON returned from Crossref into Python objects. This is done by a call to another module named doilookup.
Within the scripts, data is always handled as dictionaries with the keys in the dictionaries representing the heading of columns in a tab delimited file. The Python csv module contains two utilities called DictReader and DictWriter that read and write files to and from dictionaries. DictReader provides a fieldnames list that contains the column headers in order and DictWriter must be provided with a similar list that is used as the column headers and sets their order.
Running the Script with WOS Integration
The main script called tsv4saf [8] takes two arguments: the path and filename of the WOS input file, and the path and filename of the output file:
python3 tsv4saf.py [your input file] [your output file].
Your input file = the path to the tab-delimited file downloaded from WOS
Your output file = the desired name of your file in tsv format
For example:
python3 tsv4saf.py wosdata/savedrecs.txt foringest/output.tsv
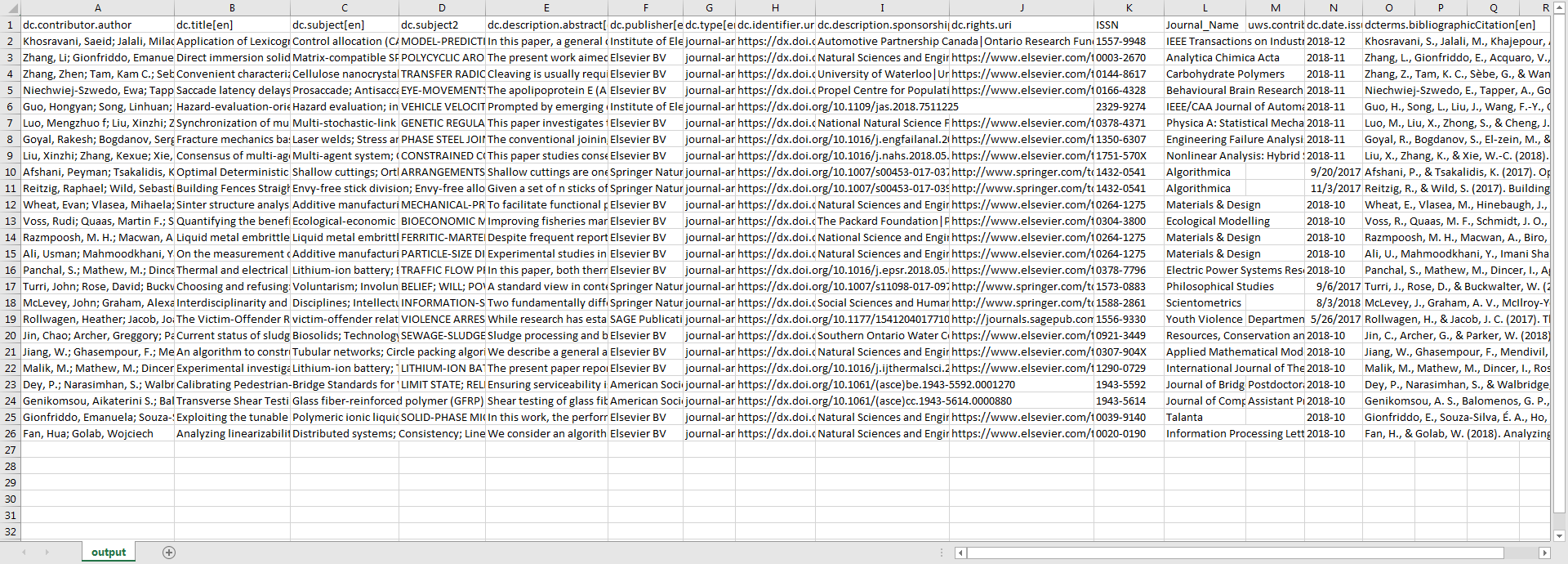
As was desired, the output of this script will return synchronized rows of metadata for the targeted Dublin Core fields, as well as some fields that are specific to local schemas and workflows.
Figure 2. Screenshot of the script’s output shown in Excel for a set of sample data. Click to enlarge
When viewing the output, it is noticeable that certain cells are returned empty. Since this script relies heavily on the existence of DOIs, if the supplied metadata from WOS is missing DOIs, the script is programmed to only output WOS metadata for those particular rows, and will skip columns normally obtained by Crossref rather than returning an error. Similarly, the script will output blank cells for data that is not existent in Crossref, as information is not always supplied.
Nonetheless, this saves significant amount of time, and helps support staff perform edits to the metadata at a much quicker rate than was possible before the implementation of the Python script.
Running the Generalized Script on a Single DOI
The Python module, tsv4saf, calls a separate module called doilookup. The doilookup module can be used by other Python scripts or used as a standalone module from the command line. This may be useful to users who are not starting with a WOS file or targeting a CSV import process for DSpace.
The module, doilookup, can be used from the command line to fetch the Crossref data for a single DOI.
python3 doilookup.py [your DOI]
When used from the command line, the script outputs a printed list of the metadata fields (using field names modeled on the Crossref JSON output) and the value found for each field (Fig. 3).
$ python3 doilookup.py 10.1002/cbic.201800049 pb: Wiley ty: journal-article id: https://dx.doi.org/10.1002/cbic.201800049 fu: Natural Sciences and Engineering Research Council of Canada|China Sponsorship Council li: http://doi.wiley.com/10.1002/tdm_license_1.1|http://onlinelibrary.wiley.com/termsAndConditions#vor issn: 1439-4227 ct: ChemBioChem af: Department of Chemistry and Waterloo Institute for Nanotechnology; University of Waterloo; 200 University Avenue West Waterloo Ontario N2L 3G1 Canada|Xiangya School of Pharmaceutical Sciences; Central South University; 172 Tongzipo Road Changsha Hunan 410013 China issued: 2018-04-26 bc: Yu, T., Zhou, W., & Liu, J. (2018). An RNA-Cleaving Catalytic DNA Accelerated by Freezing. ChemBioChem, 19(10), 1012–1017. doi:10.1002/cbic.201800049
Figure 3. Example of how to use this script on its own and the resulting output
Utilizing the doilookup for customized scripts
The doilookup script can be called from within another Python script. We have done this with our tsv4saf script which is invoked by the following method:
import doilookup
data = doilookup.lookup('[your DOI]')
Within doilookup, a function lookup() takes a DOI as its argument and returns the data in a Python dictionary. As the tsv4saf script does, this can be called within a loop to handle multiple DOIs. Assuming the variable rows is a list of dictionaries and each dictionary has a key of DI (where a DOI is stored), a new list called ‘newrows’ can be built with dictionaries that have the data from ‘rows’ together with data retrieved from Crossref:
newrows = []
for row in rows:
rowaug = doilookup.lookup(row['DI'])
newrows.append({**row, **rowaug})
The variable data will then contain a Python dictionary with the metadata in it.
The handling of the metadata within the scripts and the output of the scripts are documented by certain fixed dictionaries and lists. They are:
- in tsv4saf.py
-
- fields: a list of the fields of interest in the WOS files and in the output of the doi lookup that are used in the output
- mapping: a dictionary mapping the fields of interest to output fields for DSpace
- newfieldnames: a list of the output field names produced by applying mapping to fields
- in doilookup.py
- addedfields: a list of fields that are constructed from Crossref data
As an easy example of customization, your site will not be using our local metadata field uws.contributor.affiliation2. Your site can change the entry in mapping from:
uws.contributor.affiliation2[en]
to instead use a more standard field like:
dc.contributor
Further Considerations
The heavy lifting of parsing the Crossref JSON data is included in the doilookup script. The difficulty is in correctly parsing the JSON data returned by Crossref and finding the data of interest. This is sometimes nested within the hierarchy of the JSON data and is not always present in all records, so a certain amount of drilling down and testing of conditions is necessary. The doilookup we have provided does this for several key metadata fields of interest, but there are of course many more data options available through Crossref that could be incorporated into future versions of the script.
For those wishing to add custom fields, they should be aware that some of the JSON returned by Crossref is missing data and needs to be formatted in a way that accommodates output to CSV (or other formats). This will be the hardest part of the script to modify for people looking for different metadata and who want to format the data on output.
Those customizing the output format should be aware that our script assumes an input file that is UTF-8 encoded, and also assumes that output is done to a tab delimited file. If choosing other input and output parameters in your own project, you will need to change the parameters to the open command to read and write files or to the DictReader or DictWriter.
We also recommend that certain rules of netiquette be applied when making calls to the Crossref API [9]. This includes supplying a valid email address within your script where Crossref can reach you if any problems occur. If you wish to provide an email address, you can add a file called polite_email.txt containing your contact email. The doilookup script looks for this file and provides the address to Crossref if the file exists.
In addition, adding pauses between DOI lookups will help avoid overloading of the API. The loop within tsv4saf begins with a call to the Python sleep function to pause for a tenth of a second between loops.
Conclusion
Cory Doctorow has said, “A world of exhaustive, reliable metadata would be a utopia. It’s also a pipe-dream” (2001). As with many projects aimed at automating the collection of metadata, it’s not uncommon to encounter imperfections, and to ultimately learn that humans are still helpful and necessary intermediaries in the quality control process.
Our methodology succeeds in offering a holistic approach to how most repository staff work, and offers an attractive option for those repository managers who want to streamline a very tedious and low-level part of the otherwise challenging and rewarding work that themselves and their staff members take part in.
By using the semi-automated tsv4saf script, you can quickly collect large volumes of well-formed metadata, and have it delivered in a format that remains flexible enough to be vetted by a pair of human eyes who can inspect and improve the delivered content as needed. Further, because Python easily communicates with APIs, this method leaves open the possibility of connecting this script to many other value-added scholarly services in the process, thereby offering the potential for further enrichment of metadata, and improvements to the experiences of repository users in the long term.
Bibliography
-
- Averkamp S, Lee J. Repurposing ProQuest Metadata for Batch Ingesting ETDs into an Institutional Repository. 2009 [accessed 2018 Sep 7];(7). https://journal.code4lib.org/articles/1647
- Confederation of Open Access Repositories (COAR). Sustainable Best Practices for Populating Repositories: Preliminary Report. 2012 [accessed 2018 Sep 7]. p. 1–11.
https://www.coar-repositories.org/files/Sustainiable-practices-preliminary-results_final.pdf - Doctorow C. Metacrap: Putting the torch to seven straw-men of the meta-utopia. 2001 Aug 1 [accessed 2018 Sep 7]. https://people.well.com/user/doctorow/metacrap.htm
- Li Y. Harvesting and Repurposing Metadata from Web of Science to an Institutional Repository Using Web Services. 2016 [accessed 2018 Sep 7];22(3/4). (D-Lib Magazine). doi:10.1045/march2016-li
- Powell J, Klein M, Sompel HV de. Autoload: a pipeline for expanding the holdings of an Institutional Repository enabled by ResourceSync. 2017 [accessed 2018 Sep 7];(36). https://journal.code4lib.org/articles/12427
Notes
[1] SAFBuilder documentation is available at: https://github.com/DSpace-Labs/SAFBuilder
[2] For further information about the Digital Commons Excel Batch Upload process, see: https://vimeo.com/79930071
[3] An example of an Islandora CSV import module: https://github.com/Islandora-Collaboration-Group/icg_csv_import
[4] Scopus is likely comparable in this respect but was not tested.
[5] For brevity, we have intentionally removed certain steps that do not pertain to metadata collection such as gathering files, checking copyright, etc.
[6] Documentation (including installation) of the latest version of Habanero is available at https://habanero.readthedocs.io/en/latest/index.html
[7] For a useful discussion of DOI content negotiation through Crossref see: https://crosscite.org/docs.html
[8] GitHub Repository for both the doilookup and tsv4saf scripts: https://github.com/cpgray/tsv4saf
[9] Link to Crossref API documentation, including etiquette: https://github.com/CrossRef/rest-api-doc
About the Authors
Chris Gray is a web and database developer for the University of Waterloo Library with a strong interest in data wrangling. He prefers Python and PostgreSQL more than any ordinary brands and thinks “user-friendly” is too often used to avoid automation. He only speaks of himself in third person in special circumstances.
William Roy is Digital Repository Scholarship Specialist for the University of Waterloo library and handles recruitment, promotion and depositing of scholarly works for the institutional repository. He holds a Masters of Library and Information Science from the University of Western Ontario but is earnestly working on earning his chops on the technical side of librarianship.


Subscribe to comments: For this article | For all articles
Leave a Reply