by Colleen Fallaw, Genevieve Schmitt, Hoa Luong, Jason Colwell, and Jason Strutz
Introduction
In 2016, the University of Illinois at Urbana-Champaign (UIUC) launched Illinois Data Bank, a public access repository for publishing research data from Illinois researchers. Illinois Data Bank is part of a larger digital library system at UIUC called Medusa, which is composed of a set of internal digital preservation and management tools, along with several patron-facing access systems (Robbins, 2018). All deposited datasets receive professional curation services to ensure their completeness, understandability, and access for use in the future. In the strategic plan for 2018-2023, UIUC continues emphasizing the importance of “responsible data sharing practices throughout the institutional and constituent lifecycles” (Strategic Plan, 2018).
The year it launched, publication of the article “Overly Honest Data Repository Development” provided a holistic view of the development process for the Illinois Data Bank (Fallaw et al., 2016). The initial technical and service design choices were detailed in the article. Going into Illinois Data Bank’s fifth year of operation, researchers increasingly express a need to share large, complex datasets. This calls for reliable, highly available, cost-effective, scalable storage accessible to computation resources. The promise of these features in cloud services appeared well-suited to meet those needs, and implementation met with some meaningful success, but also with painful bumps along the way.
Challenges of Large, Complex Datasets for Ingestion, Curation, and Access
All faculty, graduate students, and staff conducting research affiliated with UIUC can deposit their research data into the Illinois Data Bank. Published datasets are then available to everyone around the world for viewing and downloading. To align with the spirit of open data for data sharing, all deposited datasets are curated to ensure completeness, understandability, and accessibility in the future. Curatorial review is a “human layer” that brings the disciplinary knowledge and software expertise necessary for reviewing incoming submissions to ensure that the data meet the FAIR principle: findability, accessibility, interoperability, and reusability (Johnston, 2018). The depth and length of curation review depends heavily on the size and structure of the dataset and how much detail is in the documentation. Based on our internal data, approximately two hours is an average amount of time spent to curate a deposited dataset in the Illinois Data Bank.
The curation services that we offer include, but are not limited to:
• Review files and metadata for completeness.
• Check documentation thoroughly for understandability.
• Validate files, including running any included computer code to the extent availability, license terms, and resources permit.
• Link to related publications and other materials.
While researchers surveyed by Springer Nature identified non-technical issues, such as “organizing data in a presentable and useful way,” as the main challenges to data sharing, file size was also found to impact data sharing. “Respondents that generate the smallest data files (<20MB; n = 2,036) have the highest proportion of data that are neither shared as supplementary information nor deposited in a repository (42%). In contrast, 70% of those with data files greater than 50GB (n = 700) share their data, with a strong preference for sharing through repositories (59%)” (Stuart et al., 2018).
Large datasets also create challenges for the curation process. In order to curate a dataset, our curator downloads as many files in the dataset as possible to check for completeness and possible errors. However, this is not always a case, especially when the dataset contains multiple large files. For example, research data from data-intensive disciplines, such as Atmospheric Sciences, Electrical, and Computer Engineering, are normally deposited in large, zipped files (some above 50GB). To curate those, the curator needs technical assistance from development and infrastructure professionals in Library IT. In most cases, physically sharing and transporting an external, portable hard drive which contains files that curator needs, would work. However, this consumes cumbersome time and resources even when participants are not navigating a pandemic, so we were motivated to find an alternative solution to not only support depositors and users of datasets, but also curators.
For this paper and in planning discussions among the staff supporting the Illinois Data bank, file size in datasets starts being “large” at the size it is not trivial to ingest and access it in a standard web form. Because our policies allow for multi-TB deposits, we have pragmatically found the need for additional solutions at approximately 4GB, 15GB, and 50GB. As shown in Figure 1, almost all the files deposited are 4GB or smaller. However, as shown in Figure 2, datasets with larger files are being published in the Illinois Data Bank with increasing frequency.
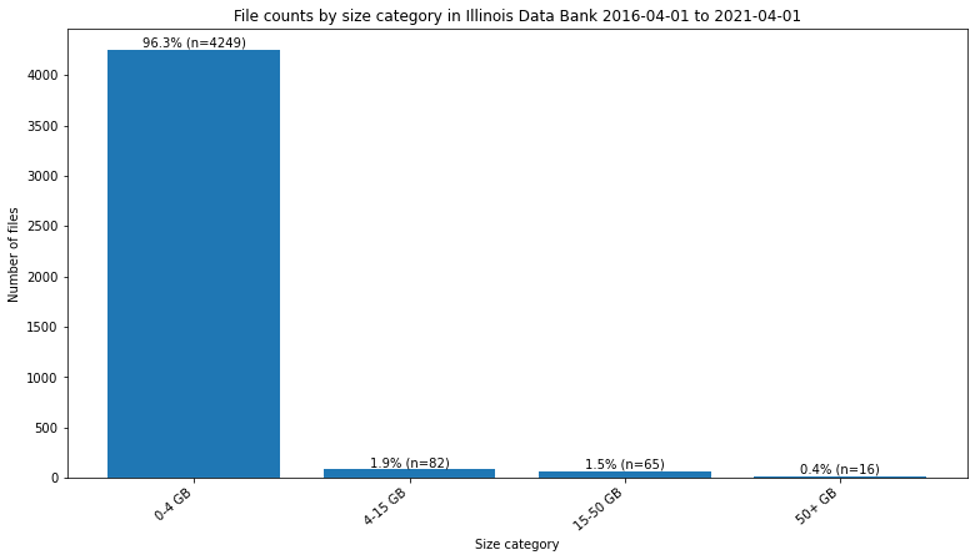
Figure 1. File counts by size category in Illinois Data Bank 2016-04-01 to 2021-04-01.
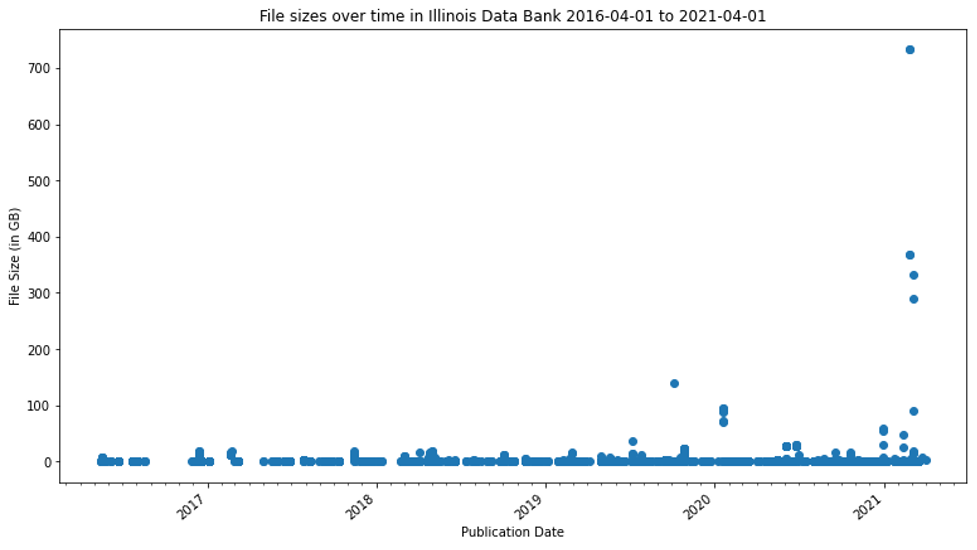
Figure 2. File sizes over time in Illinois Data Bank 2016-04-01 to 2021-04-01.
In the context of technically supporting data sharing in Illinois Data Bank, a dataset is “complex” when there is more than about one page worth of files to list in a dataset, or if the data is organized hierarchically in directories. Curators have identified archive formats such as .zip, .7z, and .tar to be preservation challenges, in part because of a lack of transparency as to the format of the files inside (Braxton, 2018). Also, in informal feedback, depositors and users of datasets in Illinois Data Bank have identified being able to see the structure of the datasets in the web application as important to data sharing. While the datafiles in Illinois Data Bank are in a large variety of file formats, 19.5% (n = 859) are in some kind of archive format, requiring additional processing to reveal the internal structure and file types for use and curation (Fallaw, 2021).
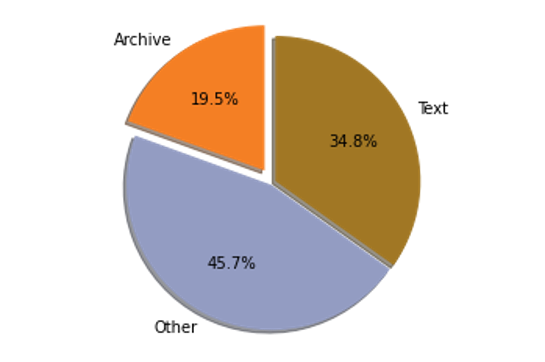
Figure 3. File type categories.
On-premises Solutions
When Illinois Data Bank launched, all storage and web service resources were hosted and administered on-campus. At launch, Illinois Data Bank supported ingestion of files up to 15 GB directly from Box through the web interface. Larger files were transferred with portable hard drives and ingested manually by technical support staff. We soon added a data file ingest API and sample python client, which worked by incrementally adding chunks to a file on a filesystem, which became the go-to solution for command-line transfer of files from research compute clusters. Box offers API access, and Illinois Data Bank uses a file import Box widget with features that include single sign-on and integrated file selection. Box has a per-file limit of 15GB. The data file ingest API was used to transfer data exclusively with command line access, which is the case for some of the compute clusters where researchers store and process data. It also supported files up to the policy limit of 2TB.
On-premises to Cloud Migration
The file storage element of Medusa at the time of Illinois Data Bank’s launch had been designed to be integrated with a compute cluster for active research projects, not to support highly-available web applications with uncertain and variable storage needs. We wanted storage designed to be more highly available, and to scale better with storage needs, which we expected to grow, though it was not clear to us how much or when. An additional element of the environment building momentum toward migration to cloud architecture was that our university’s centralized technical services group was brokering a deal with Amazon Web Services (AWS) and encouraged us to take advantage of the potential benefits they were evaluating. This aligned with trends in the library community (Goldner, 2010).
In approaching our initial migration to cloud architecture, we understood that we had a lot to learn and that every change would have potential to uncover complications, but we judged that it was worth at least a pilot program to test the concept. As is common in large innovative technical projects, it was easy to underestimate the time, effort, and complications involved. On February 18, 2019 we switched the production instances of our digital library storage and web applications from on-campus platforms to Amazon Web Services (AWS). File system storage systems are the kind of system with folders containing files like you would find on a personal computer. In contrast, AWS S3 and Google Drive are referred to as object stores. AWS does offer a flexible file system storage called Elastic File System (EFS), but it has a per-GB per-month price of ~30 cents for EFS vs ~3 cents for S3 for our initial ~150TB. We opted to adapt to S3 object storage buckets (Robbins et al., 2019).
A fully on-premises implementation was the most familiar to us. We wanted a minimum of three replicated copies in two physically separate locations in order to durably protect data, and in order to maintain a secondary read-only option during scheduled and unscheduled maintenance at the primary location. The physical, logistical, and technical challenges were daunting. Appliances that managed replicated copies were readily available on the market, but were expensive compared to bulk storage. Creating and maintaining a data center within library IT facilities would require a large initial investment with repeating refresh cycles, which could not be accommodated at the scale we needed on a consistent timeline given the fluidity of higher education budgets.
At launch, Illinois Data Bank and the supporting infrastructure was built on virtual hardware. Different functional components or software stacks were built on separate virtual servers, with CPU, RAM, disk, and network bandwidth allocated according to each function. The production environment had eight virtual servers, all of which had to be managed for operation system and software updates and monitored for security and performance limitations. The virtual cluster disk was configured for daily backup via volume snapshot, which was great for the operating system volumes but was not feasible in a realistic backup window for the amount of raw data in the repository. For that purpose, we turned to network attached storage, which replicated via rsync to a secondary block storage location in a different building. We were able to fail over by changing the mount point configurations on each application server and restarting them in the appropriate sequence.
Although the on-premises setup was cost effective, there were problems with reliability and scale. The primary and secondary storage were in different buildings from the virtual server cluster and frequently experienced network issues. An outage to any of the locations had the potential to partially or completely disable the repository. We were also required to buy additional storage in 30TB chunks with up to eight weeks of lead time due to procurement and installation delays. In order to accommodate growth and to improve stability, we decided to move all the primary repository storage to Amazon S3.
For both campus-based and cloud-based approaches, we decided to use AWS S3 Glacier as a completely separate disaster recovery location for all data. With the application and primary data on premises, we periodically uploaded an archive file (tar.gz format) with the content to be preserved to Glacier, and maintained an index of what content was in each archive. With the cloud implementation, we have cross region replication setup to copy data unidirectionally to a separate AWS region when any object is uploaded to Medusa’s S3 bucket in the primary region. This ensures strong durability at extremely low cost, with no chance of automated processes propagating accidental deletions or changes to the preservation copy. It is very slow and expensive to retrieve the entire data set, and complicated to identify a smaller part of the data set to restore in the event of disaster, but this cost was accepted as a last resort storage location for the digital objects.
Setting up mount points to S3 buckets is comparable in complexity to other types of network attached storage. However, because S3 is an object storage platform, we could take advantage of APIs to reduce or eliminate file system calls. Combined with services offered natively in AWS, such as the Relational Database Service and Elasticsearch, we were able to completely remove several virtual machines from the system. This created a shift in how infrastructure maintenance was done—instead of keeping a list of all servers that needed to be patched, monitored, and controlled for changes, we had services with monitors and alerts maintained within the AWS console. Because everything was defined in our infrastructure provisioning tool (Terraform), we could predict when changes would have an effect on other parts of the system so they could be tested, and any mistakes could be rolled back by reverting to the previous known-good configuration. We went from managing many parts of a single system separately to managing the entire system holistically.
Cloud Solutions
Some of the Medusa servers involved with supporting the Illinois Data Bank were hosted on virtual machines (VMs) administered by centralized campus IT services, while others were entirely maintained by library IT staff. While we used the infrastructure automation framework Puppet to some extent, we hoped to administer our infrastructure more efficiently and effectively—focusing on innovation rather than routine maintenance. Cloud solutions include Globus-based solutions to handle large files.
The Illinois Data Bank manages the complexity of a large number of files or hierarchically structured files by organizing them into archive files types such as zip or tar. A custom service developed in-house examined and reported on the contents of archive file types such as zip and tar to support use and curation of complex dataset directory and file structures. This service relied on filesystem tools. Our initial adaptation to cloud infrastructure involved migrating the VM to an EC2 and temporarily copying binary objects from S3s buckets to ECS. To further adapt this service to cloud infrastructure, we dove into AWS services for scalable microservices.
The differences between how file storage and object storage work required extensive adaptation of our systems and approaches to content transfer. And even file system storage provided by cloud services over the internet has connection interruptions and delays that require more flexible and robust solutions than an all on-campus infrastructure. We created custom workflows that involve the transfer of files between campus storage and cloud storage: dataset files of up to 2TB into and out of Illinois Data Bank by campus researchers.
The challenge of needing increased availability, scalability, and flexibility has been effectively addressed with our migration to cloud infrastructure. The biggest change for system administration is that library IT has full control over infrastructure. Using on campus resources relied on system administrators from other groups for provisioning and responding to incidents for storage and compute resources. Overall, library IT has a much easier time managing resources now since S3 is very reliable, and if more compute resources are needed, that can be adjusted directly. One side-effect to having full control over infrastructure is the need to better keep track of expenses.
The library IT infrastructure team was used to using monitoring tools to keep track of servers and services, but we needed a similar vision into our daily and weekly AWS infrastructure costs. After tracking expense trends in the first few months operating in AWS, we created AWS budgets to notify us when expenses are beyond what we were expecting. With the added responsibility of keeping costs under control, the change has been a net positive for the infrastructure team. It has turned many of our less fun infrastructure management tasks like provisioning and deprovisioning infrastructure, user and access management, security, and cost management into challenging puzzles using AWS’s automation tools and third-party tools like Terraform and Ansible.
While system administration became more interesting and effective, many elements of the application needed to be adapted with migration to the cloud. One of the adaptations from an all on-campus infrastructure vs. cloud object storage and web servers involved modifying the API for data transfer. There is support in the AWS S3 API for chunked file upload, although the minimum chunk size was larger than we had used previously. To adapt to cloud infrastructure and improve upload performance, library IT staff implemented tus resumable file upload protocol (tus) with S3 backend. This produced noticeably faster uploads in the web interface compared to http without the tus layer, and initial testing of the API indicated this could be a workable solution. However, “the [tus] protocol uses a data model which is mostly incompatible with AWS S3” and integrating “requires some workarounds” (Kleidl, 2016). In field conditions, our researchers started encountering frustrating reliability and transparency issues with files larger than ~100 GB using the updated API and sample client adapted to our cloud architecture. On top of the aggravation of failed transfer attempts, this led to rushed, disruptive portable hard drive transfers.
Enter Globus. As described on their website at the time of this writing, “Globus is a non-profit service for secure, reliable research data management. With Globus, subscribers can move, share, & discover data via a single interface – whether your files live on a supercomputer, lab cluster, tape archive, public cloud or your laptop, you can manage this data from anywhere, using your existing identities, via just a web browser. Developers can also use Globus to build applications and gateways leveraging our advanced identity management, single sign-on, search, authorization, and automation capabilities” (Globus, 2021).
Because Globus is a transfer service, not a separate kind of file system, it does not maintain exclusive control of the binary objects it has access to through a storage gateway. Files transferred between endpoints using Globus can be manipulated using other tools in the same location. This was key to its utility for our purpose.
Globus offers a subscription service. The University of Illinois has a campus-wide Non-Profit Research and Education subscription for Globus Online. Our subscription includes unlimited managed endpoints plus premium storage connectors for AWS S3 and Google Drive. A managed endpoint means the endpoint can use the subscription features, such as hosting shared endpoints and any premium storage connectors.
Shared endpoints (a.k.a guest collections) are required to share data using Globus Connect Server V5. Posix storage systems include the kind of system with folders containing files like you would find on a personal computer. AWS S3 and Google Drive are non-Posix object stores. Premium storage connectors are required to use non-Posix storage with an endpoint. Setting up Globus for use by Illinois Data Bank required significant organizational and technical investment initially, but it not only supported considerably smoother ingestion of data files into datasets, but library IT staff now use it for other backend services as well as additional file transfer tasks that have arisen.
Globus Ingest Steps
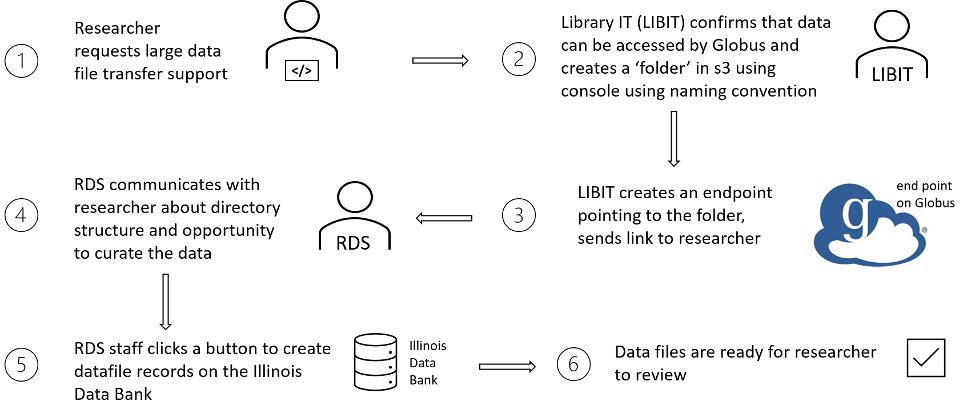
Figure 4. Globus ingest steps.
While the pain that prompted us to integrate Globus into Illinois Data Bank related to transfer in, once it was set up, we also offered transfer out, which is increasingly important as Illinois Data Bank hosts larger datasets. It also supports networked curation of large complex datasets within the Data Curation Network.
At launch, Illinois Data Bank adapted a system for supporting large downloads that was already in place for the Medusa preservation system. One of the shared backend services of Medusa is a downloader service: a Ruby on Rails application combined with an NGINX server running mod_zip and another NGINX server to allow streamable zip downloads of Medusa content. Downloads with a large number of files are served directly through Rails using a helper program clojure-zipper. Illinois Data Bank interacts with the Medusa downloader service using an API—sending a list of files and getting back a download link, which it in turn displays to the end user.
While the Medusa downloader system was updated effectively to adapt to cloud infrastructure, larger datasets can be transferred more robustly and flexibly using Globus. Also, for institutions and individuals already using Globus, some may prefer to use it when available even for smaller files.
We set up a shared Globus endpoint on an S3 bucket with read permission granted to all logged-in users. As each dataset is published, a background job copies the files to that endpoint using a naming convention. When a dataset landing page is served, Illinois Data Bank checks if the files are available in Globus. If available, the interface offers a link to the Globus File Manager page for that dataset. If problems arise from Illinois Data Bank users attempting to download very large datasets using other methods, we may disable non-Globus downloads for those datasets, where “very large” means whatever size causes problems.
Curator Preservation Challenges for Archive File Types
Many of the largest and most complex datasets include archive type files, such as .zip, .tar, or .7zip. In order to offer a listing of the contents of the archive files in the user interface and for curation analysis, we extract the files to filesystem storage and traverse the resulting tree. Other times it is possible to extract the needed information from metadata in the archive file, but even that requires tools that require the object to be stored on a filesystem.
We use S3 for long term storage instead of EFS because of the order of magnitude difference of expense mentioned above in our description of migrating from on-premises to cloud infrastructure. However, the flexibility of cloud infrastructure means that we can provision EFS on a temporary basis cost effectively when processing archive files. The process of extracting content metadata from archive files is resource intensive, but infrequent, making it a good candidate for something more flexible than a set-size VM on all the time.
To take advantage of cloud infrastructure support for flexible resource use, the archive extractor process utilizes an AWS Fargate Elastic Container Service task running a custom Docker image stored in the AWS Elastic Container Repository (ECR). The image consists of a Ruby script utilizing EFS for transient structured storage and AWS Simple Queue Service (SQS) for communication back to the Illinois Data Bank with the information extracted from the archive files. This serverless architecture is a simplification from a persistent AWS Elastic Compute Cloud (EC2) instance running a Ruby on Rails API to process the archives. The EC2 solution followed existing development paradigms during the transition from campus-hosted infrastructure to the AWS cloud. As development has settled into the AWS ecosystem it became apparent the archive extraction process could be transitioned into a serverless solution. Research archive deposits into the Illinois Data Bank are large, infrequent, and require increased compute power for short bursts of time making the extraction process ideal to utilize a serverless solution.
The most notable serverless architecture provided by AWS is Lambda. It allows developers to simply upload code as a .zip file in a variety of supported languages, seamlessly integrates with AWS Elastic File System, and is triggered through customizable JSON events. Naturally, Lambda was the initial architecture investigated for the archive extractor and after nearly a month of implementation, the limitations of this solution were brought to our attention. While extremely powerful and user-friendly for novice cloud developers, Lambda functions can only run for a maximum of 15 minutes before they timeout. Illinois Data Bank can handle files up to 2 TB which can take hours to process even under ideal computing conditions. With this constraint in mind, we worked with an AWS Technical Account Manager to brainstorm another AWS serverless solution and we decided to pursue AWS Fargate Elastic Container Service. Fargate is a serverless solution similar to Lambda with more customization and fewer restrictions which can lead to development complications. Whereas Lambda works with .zip program files, Fargate utilizes container images thus working with Fargate requires prior knowledge of Docker containers or rapid familiarization. Once the Ruby script was migrated to a Docker container and uploaded as an image, the next hurdle was connecting the Fargate task to the EFS.
Ideally, the process to set up an EFS connection with Fargate is straightforward, the file system is added to the Fargate task as a mount point and connected as an external volume. However, this relies on the AWS Virtual Private Cloud (VPC) utilizing the AWS Domain Name System (DNS), and due to university constraints, the existing VPC uses a custom DNS. In an attempt to mitigate this issue, we tried to attach the EFS volume through the Docker container itself outside of the cloud, however, this is not supported in AWS. Instead, we were left with two solutions. First, we could add DNS forwarding and tackle the setup, and maintenance while risking the stability of our existing infrastructure on the VPC or we could create a new separate VPC to use AWS DNS for the task. We chose to create a new VPC, accepting the added risk for the increased stability and reduced risk to our existing production environments on the VPC. With this new infrastructure in place, we were able to connect the EFS to the Fargate task and begin fully testing the archive extractor process.
Initial testing proved successful as smaller files were able to be uploaded to the EFS processed in the Ruby script and the output was written to the SQS to be queried and handled by the Illinois Data Bank. Subsequent testing of larger files demonstrated Fargate’s ability to compute for extended periods of time, however, it also highlighted a key limitation in the SQS communication between the Archive Extractor service and Illinois Data Bank. SQS queues have a size limit of 256 KB per message. This can be extended up to 2 GB through the SQS extended client library which is unfortunately only available for Java at the moment. This new constraint left us pondering a few different solutions including splitting JSON results into smaller messages, creating a wrapper for the Java library to be used in Ruby, or uploading the JSON result to an S3 bucket and sending the bucket key to the Illinois Data Bank as the SQS message. With a similar thought process to creating a new VPN, to minimize future maintenance and increase stability we chose to write the JSON to an S3 bucket to be read from the Illinois Data Bank backend server. This solution utilized existing AWS products without the need to maintain any new technologies ourselves. As with any technology there will always be more bugs, however, for now it appears we have worked through the major challenges of transforming an EC2 solution into a serverless architecture with minimal overhead and increased technological support via AWS.
Conclusion and Beyond
The initial technical and service design choices made at Illinois Data Bank’s launch addressed the institutional data repository challenges of depositing, using, and curating large, complex datasets with tools designed to work with file systems rather than object stores and internal networked connections between storage systems and web servers rather than cloud-based connections. During the five years since launch, library IT at UIUC has pursued improvements in reliability, performance, and flexibility through a migration of repository infrastructure to a cloud platform. We discovered and developed tools, such as Globus and Illinois Data Bank Archive Extractor. In working through what sometimes seemed like intensely nested layers of complexity and surprises in the migration, we developed new patterns of infrastructure management that afford higher levels of flexibility and position us for further innovation.
Our anticipated next targets for Institutional Data Repository development and migration efforts are using SQS to replace our RabbitMQ server, and more automation integration with Globus. We currently maintain a RabbitMQ server on an EC2 to handle communication between repository service components. We are analyzing a migration to SQS to potentially further reduce server administration and infrastructure costs. Serving as a pilot use case, the recently migrated, custom developed Archive Extractor described above uses SQS to communicate with Illinois Data Bank. While migration to SQS would be invisible to users of the systems, improved integration with Globus could improve user experience. Each step in the Globus ingest workflow represented in Figure 4 is vulnerable to staff availability, and many steps require coordination between the depositor and repository staff. Emerging features of the Globus and AWS APIs may support smoother automation of more steps. On the dataset curation and use side, very large datasets could overtax html-based download options, so limiting download options in the interface to only Globus for those datasets may be necessary for reliability and performance–for the users downloading the files as well as other users accessing repository web services at the same time.
Information sharing needs of publishers, curators, and users of datasets in institutional data repositories continue to be moving targets in an ever-shifting technology landscape. Research Data Service and Library IT staff expect to continue to use a mix of on-premises resources, cloud platforms, third party tools, custom code, and whatever comes next to meet those needs.
Acknowledgements
We thank Heidi Imker, Director of the Research Data Service and Associate Professor, University Library at UIUC, for feedback and editing.
References
[1] Braxton, S., Fallaw, C., Luong, H., Orlowska, D., Hetrick, A., Rimkus, K., Anderson, B., & Imker, H. (2018). Should we keep everything forever? Determining long-term value of research data. Poster presented at iPres2018, Boston, USA. http://hdl.handle.net/2142/91659
[2] Fallaw, C. (2021). Datafile features in Illinois Data Bank datasets 2016-04-01 to 2021-04-01. University of Illinois at Urbana-Champaign. https://doi.org/10.13012/B2IDB-7291801_V1
[3] Fallaw, C., Dunham, E., Wickes, E., Strong, D., Stein, A., Zhang, Q., Rimkus, K., Ingram, B., & Imker, H.J. (2016). Overly honest data repository development. The Code4Lib Journal, no. 34. https://journal.code4lib.org/articles/11980
[4] Gewin, V. (2016). Data sharing: An open mind on open data. Nature 529, 117–119. https://doi.org/10.1038/nj7584-117a
[5] Globus. (2021). What we do. Globus. www.globus.org/what-we-do
[6] Goldner, M. (2010). Winds of change: Libraries and cloud computing. OCLC Online Computer Library Center, Inc. https://www.oclc.org/content/dam/oclc/events/2011/files/IFLA-winds-of-change-paper.pdf
[7] Johnston, L.R., Carlson, J., Hudson-Vitale, C., Imker, H., Kozlowski, W., Olendorf, R., Stewart, C., Blake, M., Herndon, J., McGeary, T.M., Hull, E., & Coburn, E. (2018). Data curation network: A cross-institutional staffing model for curating research data. International Journal of Digital Curation, 13(1), 125-140. https://doi.org/10.2218/ijdc.v13i1.616
[8] Johnston, L. (2020). How a network of data curators can unlock the tremendous reuse value of research data. OCLC. https://blog.oclc.org/next/data-curators-network/
[9] Kim, Y., and Burns, C.S. (2016). Norms of data sharing in biological sciences: The roles of metadata, data repository, and journal and funding requirements. Journal of Information Science, 42(2), 230–245. https://doi.org/10.1177/0165551515592098
[10] Kleidl, M. (2016). Tus.io (blog). https://tus.io/blog/2016/03/07/tus-s3-backend.html
[11] Kowalczyk, S., & and Shankar, K. (2011). Data sharing in the sciences. Annual Review of Information Science and Technology Journal, 45, 247-294. https://doi.org/10.1002/aris.2011.1440450113
[12] National Science Board. (2011). Digital research data sharing and management. https://www.nsf.gov/nsb/publications/2011/nsb1124.pdf
[13] Office of Science and Technology Policy. https://web.archive.org/web/20160304043850/https:/www.whitehouse.gov/sites/default/files/microsites/ostp/ostp_public_access_memo_2013.pdf
[14] Office of the Provost. (n.d.). The Illinois strategic plan, The University of Illinois at Urbana-Champaign. https://strategicplan.illinois.edu/2013-2016/goals.html
[15] Office of the Provost. (n.d.). The next 150: Strategic plan for 2018-2023. The University of Illinois at Urbana-Champaign. https://strategicplan.illinois.edu/
[16]Robbins, S., Troy, J., Rimkus, K., Strutz, W.J., & Colwell, J. (2019). The challenges and charms of a cloud-based repository infrastructure transition: Lifting and shifting the library’s Medusa repository. Presented at Open Repositories 2019 Conference, Hamburg, June 13, 2019. http://hdl.handle.net/2142/104029
[17] Robbins, S. (2018). Medusa: Service-oriented repository architecture at the University of Illinois. Presented at Open Repositories 2018 Conference, Bozeman, MT, June 6, 2018. http://hdl.handle.net/2142/100077
[18] Strutz, J. (2017). The Medusa repository: Turning data into stone. Presented at The University of Illinois at Urbana-Champaign IT Pro Forum November 1, 2017
[19] Stuart, D., Baynes, G., Hrynaszkiewicz, I., Allin, K., Penny, D., Lucraft, M., & Astell, M. (2018). Whitepaper: Practical challenges for researchers in data sharing. https://doi.org/10.6084/m9.figshare.5975011.v1
[20] Tenopir, C., Dalton, E.D., Allard, S., Frame, M., Pjesivac, I., Birch, B., Pollock, D. & Dorsett, K. (2015). Changes in data sharing and data reuse practices and perceptions among scientists worldwide. PLOS ONE, 10(8): e0134826. https://doi.org/10.1371/journal.pone.0134826
[21] Tenopir, C., Allard, S., Douglass K., Aydinoglu, A.U., Wu, L., Read, E., Manoff, M., & Frame, M. (2011). “Data sharing by scientists: Practices and perceptions.” PLOS ONE 6,(6): e21101. https://doi.org/10.1371/journal.pone.0021101
About the Authors
Colleen Fallaw
Research Programmer at the University of Illinois at Urbana-Champaign Library
http://orcid.org/0000-0002-0339-9809
mfall3@illinois.edu
Genevieve Schmitt
Software Engineer at the University of Illinois at Urbana-Champaign Library
https://orcid.org/0000-0002-4974-9501
gschmitt@illinois.edu
Hoa Luong
Associate Director, Research Data Service at the University of Illinois at Urbana-Champaign Library
https://orcid.org/0000-0001-6758-5419
hluong2@illinois.edu
Jason Colwell
Systems Administrator at the University of Illinois at Urbana-Champaign Library
https://orcid.org/0000-0003-4150-9172
colwell3@illinois.edu
W. Jason Strutz
Manager, IT Infrastructure at the University of Illinois at Urbana-Champaign Library
https://orcid.org/0000-0002-0284-3517
strutz@illinois.edu


Subscribe to comments: For this article | For all articles
Leave a Reply