by Connor B. Bailey, Fedor F. Balakirev, and Lyudmila L. Balakireva
Introduction
With the increased prevalence of machine learning technology, the viability of big data projects, and the rapid proliferation of vast quantities of information, data are becoming more usable and useful than ever. However, these datasets must be properly stored and managed to ensure that the greatest possible scientific output can be gained from them. To achieve these goals, there is a push to use more accessible data standards to maximize the findability, accessibility, interoperability, and reusability (FAIR) of scientific data (Wilkinson 2016). Open data repositories help with the adoption and usage of the FAIR principles by providing storage for scientific data repositories and act as hubs for sharing such information. TDMS [1] and HDF5 [2] are hierarchical data formats that are commonly used in scientific communities, are well supported by and compatible with popular scientific software, and offer a wide variety of benefits over alternative data storage methods. The hierarchical design allows for many different ways of structuring and organizing data, including large and complex datasets, while also allowing metadata to be included at various levels in the hierarchy, which can help explain the structure, contents, and format of the data. Having metadata embedded within the hierarchy of the file can improve its reusability by providing inseparable, detailed information about the various parts and contents of the file. The adoption of FAIR data practices can be greatly enhanced by hierarchical data storage formats as there are several overlaps between the benefits and use cases of each tool. We found, however, a disconnect between hierarchical data formats and open data repositories and control systems that could be addressed.
The Advantages of Hierarchical Data Storage Formats
There are many reasons to use a hierarchical data storage method and many advantages if that storage type is used. These storage types allow for vast quantities of data to be organized efficiently to store arrays of data and the associated metadata that contextualize them.
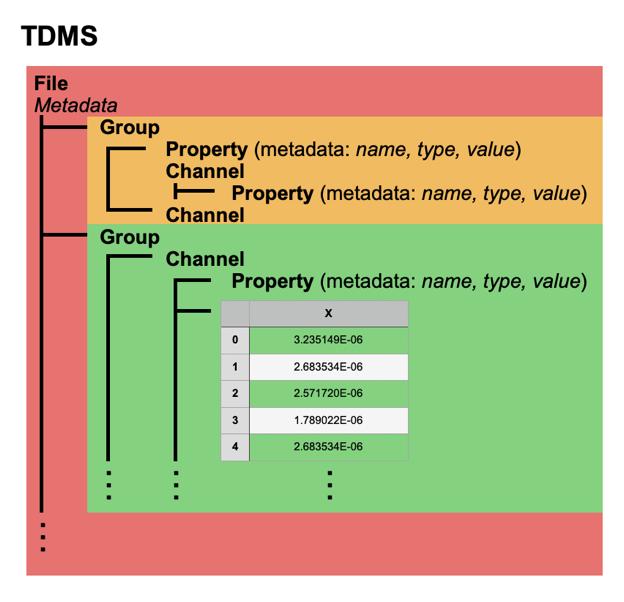
Figure 1 TDMS Sample File Structure: TDMS files consist of a three level hierarchy: file, group, and channel and can contain two types of data: property metadata and data values. Every TDMS object can have an unlimited number of properties consisting of a combination of a name (always a string), a type identifier, and a value. Channel objects can contain raw data multi-dimensional arrays.
Hierarchical data storage formats like TDMS and HDF5 allow the user to organize data arrays into groups and trees, separating out different trials or conditions, and create any grouping that helps the data make more sense. Additionally, important information such as the parameters that correspond to that dataset can be included at the same level as the data embedded within the hierarchy of the storage method. Important information such as how the data should be read can also be included at this level; for instance, waveform or time interval data that is integral to the understanding of the data channel. Because hierarchical formats work well for time series data, it is a useful format for signal processing, network monitoring, finance, weather forecasting, and largely in any domain of applied science and engineering.
The Advancement of FAIR Data Principles
The concept of FAIR was introduced in “The fair guiding principles for scientific data management and stewardship” by Wilkinson et al. in 2016 and is seen as a consolidation of good data stewardship practices (Jacobsen et al. 2020). Since then these guidelines have seen success in impacting institutions by pushing them towards using better management methods for their data. Built into the acronym are the four core goals of FAIR that institutions should strive for with their data: findability, accessibility, interoperability, and reuse. Each has significant importance; however, they all aim to reduce the chance that some valuable data can be lost to future scientific endeavors due to poor data management. Each goal is also further detailed in the guidelines and applies to any digital object being generated or stored within a repository. The FAIR principles do not dictate any kind of specific software or service, however many data repositories exist which can help institutions to adopt the principles (Wilkinson et al. 2016). Open Science Framework (OSF) is one such data repository that seeks to facilitate open collaboration in scientific research run by the Center for Open Science (COS), a non-profit technology organization. Researchers can use OSF to research and find data related to their work, and they can use it to host the data they produce themselves. These types of repositories seek to be a hub for data management, helping to facilitate the exchange of valuable scientific data between researchers.
An important part of FAIR is the metadata associated with data sets. Metadata gives important information about data; identifying it, contextualizing it, and ensuring that the full meaning of the data can be understood. The globally unique, persistent identifier that is required by the FAIR guidelines, for example, is an extremely important bit of metadata, and references to other identifiers are useful too. Many of the ideas in the FAIR principles relate to “machine-actionability”, however there is an important human interactivity element to FAIR which we focus on with our tool here. We found that FAIR data repositories, even the most well-known, like Harvard Dataverse [3] or OSF [4], lack the support of TDMS and HDF file viewing. This deters scientists from engaging with FAIR practices like publishing datasets for reuse in the open. The repositories store hierarchical format as a binary blob, so users can not see the experiment content of the file or associated metadata. This creates the problem that even if one researcher posts the TDMS file to the repository those files are not browsable or findable by others in the scientific community. We created a renderer tool to address the deficiency.
Many files on OSF can be viewed directly in the browser while looking through the repository. However, many types of files may display as plaintext, spreadsheet, or pdf style renders which may not be able to show the full complexity of data. Many files such as TDMS and HDF5 do not display at all. A rich view of data and metadata would allow a researcher to get a more complete understanding of the data they are viewing directly in the data repository.
The Hierarchical Renderer for FAIR Repositories
OSF [5] is an open source project and the rendering is done by a tool called the modular file renderer (MFR) [6]. The MFR is made up of “handlers” which handle web requests made to the MFR, “providers” which know how to take a url for a file and retrieve the necessary content for the file, and “extensions” which generate the necessary HTML for the rendering of the file. OSF allows us to integrate new “extensions” into the existing MFR framework which are called to handle the rendering of hierarchical data files. We have consulted with the staff at COS [7] so that our renderer will be added to the main OSF library and be usable by all users of OSF. Using the nptdms [8] and h5py [9] libraries in Python, the renderer extracts the data from the hierarchical data files into the Python environment and generates the necessary HTML, Javascript, and CSS content to display the rendering in the browser. In order to create this tool, we created a local development environment and ran a local host of OSF in a Docker instance. [10] The same steps should be taken to test the tool before it has been implemented in the main OSF distribution. Using these tools we were able to fork the MFR, develop hierarchical data extensions, and test its functionality locally. The main difficulties arising from a development project like this come from getting the wide variety of disparate software and libraries to properly work together.
The renderer tool [11] that we have created allows users to fully render a view of a hierarchical data file directly in a FAIR data repository. Specifically, we can render TDMS and HDF5 files in the OSF file viewer. This allows researchers and others using OSF to store their TDMS and HDF5 files in that repository knowing that it will have full support when browsing their files in the future. Our renderer builds off of tools such as the NI TDMS Viewer, a labview program that loads and displays TDMS files within a LabVIEW program. Our renderer gives users the same information directly in the browser while interacting with the files in the OSF repository. It is able to display the metadata alongside a table of the data and a plot of the data, visualizing the logical link between elements of data and metadata in a composite view. This allows us to quickly render the necessary information for someone who is browsing through these types of files.

Figure 2 TDMS Renderer Rendering a Sample File: The renderer presents the file content in a split view with the metadata and properties values being shown in tree and table formats on the left and the channel data being displayed in a plot and table format on the right. The metadata tree is expandable, collapsible, and scrollable so that the necessary information is as easy to access as possible.
The renderer view is split vertically between the tree style metadata display on the left and the data plot and table on the right (Fig. 2). On the left, in an intuitive hierarchical tree structure, the file level metadata is displayed at the top followed by the group and channel metadata and properties table beneath it. Each level is expandable, collapsible, and scrollable to allow easy access to whatever level the user is interested in looking at more closely. On the right, an image of the channel data arrays plotted and exported using the matplotlib Python library [12] is displayed. This is a quick and efficient way of rendering a plot of the data so that a user can see at a glance what the structure of the data looks like. Because the renderer isn’t designed for in depth data analysis, we found this method to be the best way to achieve fast rendering speed while also giving a visual indication of the files contents. A chunk of the data is also included in a data table beneath the plot. The table loads only the first 100 entries in a channel, again to allow for speedy load times, while still giving the user the ability to see what the channel fields of the file are and what the data they contain looks like.

Figure 3 TDMS Renderer within the OSF File Viewer: The view a user gets when accessing a TDMS file within the web browser accessing the OSF repository. The OSF toolbar is displayed at the top of the screen, the file is displayed in a toolbar on the left side of the screen and the main rendering view is shown in the right center of the screen. Selecting a TDMS file from the file menu on the left calls on our rendering tool to render it in the webpage.
The person viewing these files is likely to want to get a general idea of the file they are looking at directly in the OSF data repository. They can then identify files they may want to work with further and download those specific files to do more in depth analysis or calculations with.
Conclusion
Our rendering tool seeks to target a direct need in the scientific community not only to give researchers better tools for accessing and viewing their data, but to ease the adoption of better methods of data management. Our solution aids in bridging the gap between hierarchical data storage techniques and FAIR data repositories, making both of them more viable options. By bringing this user-friendly tool to a popular FAIR data repository, we hope that we have helped bridge that gap. Scientific institutions like LANL which have been put off by the lack of integration between them now know they can use these tools together and use them more efficiently and effectively.
Acknowledgments
The National High Magnetic Field Laboratory is supported by the National Science Foundation through NSF/DMR-1644779, the State of Florida, and the U.S. Department of Energy.
References
Wilkinson, M. Dumontier, I. Aalbersberg, G. Appleton, M. Axton, A. Baak, et al. 2016. The fair guiding principles for scientific data management and stewardship. Scientific Data 3(Mar).
Jacobsen, R. deMiranda, Azevedo, N.Juty, D.Batista, S.Coles,R.Cornet, et al. 2020. Fair principles: Interpretations and implementation considerations. Data Intelligence 2(1-2):10–29.
Endnotes
[1] TDMS file format description and documentation from National Instruments: https://www.ni.com/en-us/support/documentation/supplemental/06/the-ni-tdms-file-format.html
[2] HDF5 file format description and documentation from HDF Group: https://portal.hdfgroup.org/display/HDF5/HDF5
[3] Harvard Dataverse repository: https://dataverse.harvard.edu
[4] Open Science Framework repository: https://osf.io
[5] Open source development of osf.io, the OSF repository on GitHub: https://github.com/CenterForOpenScience/osf.io
[6] Open Source development of the modular file renderer for OSF on GitHub: https://github.com/CenterForOpenScience/modular-file-renderer
[7] Acknowledgements to Fitz Elliot at COS for consultation on OSF and MFR
[8] Documentation for the nptdms library that provides interfacing between TDMS and Python: https://nptdms.readthedocs.io/en/stable/index.html
[9] Documentation for the h5py library that provides interfacing between HDF5 and Python: https://www.h5py.org
[10] Guide to setting up a local development environment for OSF allowing you to run the extended version including our renderer until it is published on the main OSF repository: https://github.com/cbb-lanl/modular-file-renderer/blob/develop/configuringDevEnv.md
[11] Fork of the modular file renderer containing our additional extensions in the mfr/extensions folder: https://github.com/cbb-lanl/modular-file-renderer
[12] Documentation for the matplotlib library which provides plotting functions in Python: https://matplotlib.org
About the Authors
Connor Bailey (cbbcbail@gmail.com) is a graduate student at the Pulsed Field Facility, National High Magnetic Field Laboratory, Los Alamos National Laboratory, USA (MAGLAB). Connor works on simulation and visualization strategies for radio transmissions and develops automated data capture, processing, and software programming tools at the MAGLAB user facility.
Fedor Balakirev (fedor@lanl.gov) is a staff scientist at the Pulsed Field Facility, National High Magnetic Field Laboratory, Los Alamos National Laboratory, USA. The focus of his research is studies of materials under extreme conditions of high magnetic fields, including the development of the measurement infrastructure, instrumentation, and computing suites for the MAGLAB. Fedor holds a Ph.D. in Physics from Rutgers University, USA.
Lyudmila Balakireva (ludab@lanl.gov) is a research engineer of the Prototyping Team at the Los Alamos National Laboratory Research Library. In this role, she focuses on research and development efforts in the realm of web archiving, scholarly communication, digital system interoperability, repositories, and data management. Lyudmila holds a Ph.D. in Mathematics and Physics from Moscow Institute of Physics and Technology, Russia.


Subscribe to comments: For this article | For all articles
Leave a Reply