by Jennifer Ye Moon-Chung
1. Introduction
The Gumberg Library at Duquesne University has been utilizing the ILLiad statistics tool from ATLAS Systems with support from OCLC. This software not only facilitates efficient Interlibrary Loan (ILL) transactions but also provides valuable statistical data and web-based reporting systems (Atlas Systems 2019). Through the analysis of the data gathered via ILLiad, Gumberg Library has been able to review monthly trends in request rates, fulfillment rates (the percentage of successfully filled requests), and reasons for request cancellations, considering both lending and borrowing activities involving returnable items, such as physical books, and non-returnable items like electronic journal articles and book chapters. These metrics have proven instrumental in understanding the demand for different document types, analyzing usage patterns by date, and identifying the specific school or program within our institution making the most requests. Moreover, the statistics derived from ILLiad have played a vital role in completing the library’s annual report, offering a comprehensive overview of ILL transactions (ACRL 2006). This data can be accessed through a monitoring tool managed by Gumberg Library known as the “ILLiad Activity” Dashboard. [1]
However, the Gumberg Library sought to delve deeper and leverage this data to make informed decisions regarding journal subscriptions, as these subscriptions represent a significant portion of the library’s budget (Petrick 2002). One noteworthy observation from the analysis of the “Reasons for Cancellations” data, accessible through the “ILLiad Activity” dashboard, was the significant number of cancellations resulting from fulfilling requests through existing locally-held journal subscriptions. This observation prompted the library to critically evaluate its subscription decisions, considering whether to add new journals or retain existing ones, with a focus on data-driven analysis.
Developing a new reporting system to address this question posed certain difficulties. The data required for analysis was primarily created by patrons who requested Interlibrary Loan materials. While the library provides a form for patrons to fill out, there are no additional steps or verification process involved between the form and the data entered by patrons is stored directly in the ILLiad system. Consequently, the library faced numerous typos, inconsistencies, and errors in the data when downloading reports from the ILLiad reporting system. Additionally, the aggregated data offered by ILLiad did not fully align with the library’s specific reporting needs, necessitating customization. For instance, although we could obtain information about the most requested loans (limited to title information) within a selected date range throughout the ILLiad web reporting system, we desired additional insights such as whether those specific title requests were filled, cancelled, and the reasons for request cancellations if applicable. Furthermore, due to concerns regarding personally identifiable information, we remove entire transaction records from the system after a specified period of time.
To address these challenges, we utilized custom Python code. While Python provides a wide range of open-source libraries, we opted for simplicity in this specific task and relied on a minimal set of libraries, including pandas [2] for data manipulation and cleaning tasks, as well as requests [3] and json [4] for efficient communication through API calls via HTTP, ensuring effective data retrieval for analysis purposes.
The code presented in this paper has been specifically developed to address the unique requirements of processing collected Journal Request data. While the term “customized” implies that its direct applicability may vary in other scenarios, sharing this information aims to offer real-world use cases and assist others in overcoming similar challenges. By adapting the code to their specific needs, making necessary adjustments, and exploring innovative solutions, practitioners can collectively make progress in effectively handling issues associated with messy data.
2. Methods
2-1. Importing Libraries and Accessing Dataset
To begin the customized code implementation, the first step involves importing the required libraries or modules. In the presented code snippet, the following libraries are imported: pandas, datetime, json, and requests.
import pandas as pd from datetime import datetime import json import requests
Next, the code accesses the library staff’s designated box.com folder, which serves as the storage location for the monthly harvest data from the ILLiad system. [5] To ensure the security of Duquesne online services, which are protected by single sign-on (SSO) authentication, we utilized box.com. If library staff have their Box folder, or any shared drive (e.g., OneDrive or Google Drive) synced to their desktops, the default file paths for Windows are as follows:
C:\Users\{username}\Box>
C:\Users\{username}\OneDrive>
C:\Users\{username}\Google Drive>
The term {multipass_id} is used interchangeably with {username}, as in the Duquesne environment they are identical. By prompting the library staff to enter their Multipass ID, the input file path is programmatically constructed, preventing accidental modifications to the code and ensuring exclusive access to the Box folder by authorized staff.
# Note: Please ensure that the inputted data is in Excel format(.xlsx).
multipass_id = input("Enter your multipass ID: ")
input_name = input("Enter ILLiad Monthly File Name: `Do not enter .xlsx part`" )
input_file_name = input_name+".xlsx"
input_file_path = (f"C:\\Users\\{multipass_id}\\Box\\\Annual Report Procedures\\ILLiad Statistics\\ill_titles_data\\{input_file_name}")
excel_object = pd.ExcelFile(input_file_path)
df_all = pd.read_excel(input_file_path, sheet_name = excel_object.sheet_names[0])
df_all.info()
Please note that this code is specifically designed to work with Excel files (.xlsx) since we export ILLiad data in this format on a monthly basis. Library staff can modify the line input_file_name = input_name+".xlsx" to input_file_name = input_name+".csv" if they wish to use CSV files by utilizing with the pd.read_csv [6] method instead of pd.read_excel [7]. We have tested this code on Windows 10 with Python version 3.8.13 and higher, but library staff should ensure compatibility with their Python environment for optimal results.
Furthermore, to use this code, library staff need to modify the input_file_path variable with the file directory where their data is stored. For Mac OS users, the file path can be adjusted as follows:
input_file_path =(f"/Users/{multipass_id}/Box/Annual Report Procedures/ILLiad Statistics/ill_titles_data/{input_file_name}")
This structure points directly to the file directory. Therefore, if the file is stored in the Downloads folder, the path would be Users/{user_name}/Downloads/{input_file_name}. Alternatively, instead of pointing to a local file, the input_file_path can also be set to open a URL, such as input_file_path = "https://github.com/imyem7/duq_open/raw/main/sample_data.xlsx".[8]
Enter your multipass ID: my_username Enter ILLiad Monthly File Name: `Do not enter .xlsx part`: illiad-export-april-2020 <class 'pandas.core.frame.DataFrame'> RangeIndex: 1674 entries, 0 to 1673 Columns: 219 entries, Transaction Number to Courier Info dtypes: bool(1), datetime64[ns](4), float64(90), int64(2), object(122) memory usage: 2.8+ MB
Figure 1. Code Execution Result − Summary of Raw Data, “illiad-export-april-2020”
The test dataset used as a sample is the April 2020 ILLiad usage data, deliberately selected due to its relevance during the COVID-19 pandemic when interlibrary loan transactions were at their highest, effectively demonstrating the impact of this tool. This process can be applied to any current dataset and is currently employed as part of the dashboard update procedure.
The dataset originally contained 1674 rows and 219 columns, including sensitive patron information that necessitates careful handling, as is common with many usage datasets (Figure 1). However, since patron information is not required for analyzing the ILLiad journal request report, we sliced the dataset to remove unnecessary and potentially sensitive data by selecting only the columns necessary effectively anonymizing the data. The following code block demonstrates how to select the necessary data when defining a data frame:
ill_df = pd.read_excel(input_file_path, usecols=["Transaction Number",\
"Request Type", "Process Type", "Photo Journal Title",\
"Photo Journal Year", "ISSN", "Creation Date", "Status",\
"Transaction Status", "Reason For Cancellation", "Document Type",\
"Department"])
# Filter for only Article requests
filter = ill_df["Request Type"].isin(["Article"])
article = ill_df[filter]
article.info()
Required columns are selected using the usecols parameter, which includes “Transaction Number”, “Request Type”,”Process Type”, “Photo Journal Title”, “Photo Journal Year”, “ISSN”, “Creation Date”,”Status”, “Transaction Status”, “Reason For Cancellation”, “Document Type”, “Department”. The dataset is further filtered to include only the rows where the request type is “Article” using the pandas DataFrame.isin() method.
<class 'pandas.core.frame.DataFrame'> Int64Index: 1629 entries, 1 to 1673 Data columns (total 12 columns): # Column Non-Null Count Dtype --- ------ -------------- ----- 0 Transaction Number 1629 non-null int64 1 Request Type 1629 non-null object 2 Photo Journal Title 1625 non-null object 3 Photo Journal Year 1589 non-null object 4 Transaction Status 1629 non-null object 5 ISSN 1597 non-null object 6 Reason For Cancellation 767 non-null object 7 Process Type 1629 non-null object 8 Document Type 1172 non-null object 9 Creation Date 1629 non-null datetime64[ns] 10 Status 1629 non-null object 11 Department 1626 non-null object dtypes: datetime64[ns](1), int64(1), object(10) memory usage: 165.4+ KB
Figure 2. Code Execution Result − Summary of Sliced Data, a Subset of “illiad-export-april-2020”
As a result, the dataset is reduced from 1674 rows and 219 columns (Figure 1) to 1629 rows and 12 columns (Figure 2).
2.2. String Manipulation
String manipulation techniques played a crucial role in this project. They typically involve a variety of methods, including removing white spaces, finding and replacing specific characters, changing case, splitting strings into substrings, and joining multiple strings together. Our primary focus in this project was on cleaning the ISSNs and International Standard Book Numbers (ISBNs) within the ILLiad system which had been directly input by users. Although we didn’t require it for this particular project, in more complex cases, regular expressions using the re module would be worth considering. [9]
The ISSN typically consists of two groups of four digits, separated by a hyphen [-] following the format “ISSN 0123-4567”. However, users often input ISSNs with variations, such as removing hyphens, leaving spaces, or omitting spaces altogether, which complicates the automation process for practitioners. Similarly, ISBNs present additional complexities. With the transition of the ISBN system from ten-digit to thirteen-digit ISBNs since 2007, and the inclusion of hyphens in both formats, the range of variations in ISBN entry becomes even wider. The provided code result includes a randomly selected dataset from rows 330 to 350, showing the ISSN and ISBN variations along with their respective frequencies (Figure 3).
1538-9235; 1040-5488 1 0015-5357 1 0361-5227 1 9780761922605 1 9781555811044 1 9780890891278 1 9789004122581 1 0250 6262 1 1041-6080 1 0022-2224 1 1439-9598 1 1066-5684 1 978-0-7456-6130-8 1 0147-9563 1 1387-6740 1 0039-7911 1 2397-334X 1 0079-6123 1 0882-8245 1 0074-7742 1 Name: ISSN, dtype: int64
Figure 3. Variations in ISSN and ISBN Entries
In this study, our approach involved removing all white spaces and hyphens from the ISSN numbers using the strip() and replace() methods. For instance, df.replace("-", "") replaced hyphens with an empty string, and replace(" ", "") removed the white spaces between numbers. The strip() method was used to remove leading and trailing characters, including empty spaces. The code below provides an example of the approach we implemented.
issn = '1234-5678'
issn_clean = issn.replace("-", "").replace(" ","").strip()
print(issn_clean)
issn = '1234 5678'
issn_clean = issn.replace("-", "").replace(" ","").strip()
print(issn_clean)
issn = ' 1234-5678'
issn_clean = issn.replace("-", "").replace(" ","").strip()
print(issn_clean)
While ISSN numbers may vary in format, the resulting cleaned ISSN (“issn_clean”) is consistently represented as “12345678” (Figure 4).
12345678 12345678 12345678
Figure 4. Code Execution Result − The Numeric ISSN
Once we obtained the numeric ISSN digits, which have been cleaned of symbols or spaces (regardless of whether they were initially properly placed), we reinserted the hyphen (-) in the appropriate place. This step was necessary because most ISSN databases utilize hyphenated ISSNs and expect the correct formatting. The following code block demonstrates how to format an ISSN number appropriately.
issn=str("12345678")
x_issn= issn[0:4] + "-" + issn[4:8]
As a result, the output value “12345678” (Figure 5) becomes “1234-5678” (Figure 5).
'1234-5678'
Figure 5. Code Execution Result − Formatted ISSN
2.3. API Requests
Unlike cleaning ISSN or ISBN entries, cleaning the title information entered by users can be a more complex task. We have observed a variety of variations, including mixed use of upper and lower cases, incorrect spacing, typos, and incomplete titles. While advanced string manipulation techniques, such as using regular expression, can enhance the accuracy, limitations still exist, particularly when title entry itself is incomplete. Instead of directly cleaning the title entries, we opted to verify title information based on the ISSN or ISBN data. Our approach involved retrieving the journal or book title from the selected databases using API requests.
In the context of this project, while we recognized that there are other potential data sources available, we chose CrossRef and Google Books as our data sources based on several factors. These include the robust documentation provided by these platforms, the availability of accurate and extensive data, and the convenience of free access.
CrossRef is a widely recognized database in the academic community, primarily focused on scholarly content, including journal articles and conference papers. By leveraging the CrossRef REST API, we can access a vast collection of bibliographic data, such as publication titles, authors, abstracts, and even full-text articles in some cases. [10] To retrieve metadata from CrossRef, we utilized the simple requests library, which allows sending HTTP/1.1 requests. The data received from the API is in JSON format, which can be parsed using the json method. Python users also have the option to use third-party libraries for interacting with CrossRef, such as crossref-commons [11], habanero [12], crossrefapi [13].
Once we chose the library to be utilized, the initial step was to identify the suitable endpoints that expose the specific information required. This information can be found on the Crossref Unified Resource API page (https://api.crossref.org/swagger-ui/index.html). In our case, we utilized the "/Journals/{issn}" endpoint to retrieve journal information based on an ISSN. By constructing the proper URL, we could make a request and receive a response containing the desired data. For example, to retrieve journal information for the ISSN “1940-5758”, we defined the “issn” variable as “1940-5758” and the “api_url” variable as “f’https://api.crossref.org/journals/{issn}'”. We then initiated a GET request using the requests library and stored the response in the “response” variable. Finally, we were able to access and parse the data in JSON format using the response.json() method. Here is the corresponding code we tested:
issn = "1940-5758"
api_url = f"https://api.crossref.org/journals/{issn}"
headers = { 'User-Agent': 'ISSN-info/1.0 (mailto:myemail@email.com)'}
# insert your project name and contact info
response = requests.get(api_url, headers = headers)
data = response.json()
data
By executing the code, metadata for the journal with the ISSN “1940-5758” was retrieved (Figure 6). The parsed data included various details, such as the journal’s title, which in this case was “code4lib”.
{'status': 'ok',
'message-type': 'journal',
'message-version': '1.0.0',
'message': {'last-status-check-time': 1687724240530,
'counts': {'current-dois': 1, 'backfile-dois': 3, 'total-dois': 4},
'breakdowns': {'dois-by-issued-year': [[2018, 2], [2021, 1], [2019, 1]]},
'publisher': 'Crossref Test',
'coverage': {'affiliations-current': 0.0,
…
Some results omitted for space limitations.
…
'title': 'code4lib',
….
Some results omitted for space limitations.
'ISSN': ['1940-5758'],
'issn-type': [{'value': '1940-5758', 'type': 'print'}]}}
Figure 6. Code Execution Result − Journal Information Associated with the ISSN “1940-5758”
Similarly, the Google Books API provides access to Google Books’ extensive database, allowing us to retrieve comprehensive book information such as titles, authors, publishers, descriptions, and more [14]. To initiate an API call, we followed a similar process as with the CrossRef API, but with one major difference: authentication is required for Google API calls, as specified in the Google API terms of use. To test this process, users are required to obtain an API key from the Google Cloud Console (https://console.cloud.google.com/). Once authentication is completed, we constructed the API URL by combining the base URL for Google Books API (https://www.googleapis.com/books/v1/volumes), the query parameter specifying the ISBN to search for (q=isbn:{isbn}), and the API key (key={api_key}). For example, if we wanted to retrieve metadata associated with the ISBN “9781530051120”, the code would be as follows:
isbn= "9781530051120"
api_key = "-----------------" # insert Google API key
url = "https://www.googleapis.com/books/v1/volumes?q=isbn:" + isbn\
+ "&key=" + api_key
response = requests.get(url)
book_data = response.json()
book_data
As a result, we were able to confirm that the book associated with the ISBN “9781530051120” is titled “Python for Everybody,” along with other relevant information (Figure 7).
{'kind': 'books#volumes',
'totalItems': 1,
'items': [{'kind': 'books#volume',
'id': 'zjqzDAEACAAJ',
'etag': 'O2x+zybqctM',
'selfLink': 'https://www.googleapis.com/books/v1/volumes/zjqzDAEACAAJ',
'volumeInfo': {'title': 'Python for Everybody',
'subtitle': 'Exploring Data in Python 3',
'authors': ['Charles R. Severance'],
'publishedDate': '2016-04-09',
'description': 'Python for Everybody is designed to introduce
'industryIdentifiers': [{'type': 'ISBN_10', 'identifier': '1530051126'},
{'type': 'ISBN_13', 'identifier': '9781530051120'}],
'readingModes': {'text': False, 'image': False},
'pageCount': 242,
'printType': 'BOOK',
….
Some results omitted for space limitations.
…
Figure 7. Code Execution Result − Metadata associated with the ISBN “9781530051120”
Furthermore, by understanding the JSON structure of the response, we were able to extract the specific information we needed. In this case, we observed that the result data is structured within nested curly braces,{}. The “title” information is located under “volumeInfo”, which, in turn, is nested under “items”. The following code demonstrates how we extracted the title information.
items = book_data.get('items', [])
volume_info = items[0].get('volumeInfo', {})
title = volume_info.get('title')
print("Title:", title)
The result of our extraction process was as follows:
Title: Python for Everybody
Figure 8. Code Execution Result − Extracted Book Title from Nested JSON Structure
2.4. Automating the Process with a for loop
In order to automate the process, considering the limitations in library data work such as limited staff and time constraints, we utilized a for loop in Python. The for loop operates by iterating through a provided list of items and executing the defined steps sequentially for each item. For instance, we conducted a test using a list of ISSN numbers, such as "194057-58", "1940.5758", and " 19405758". The for loop iterated through this list, performing tasks such as cleaning the ISSN numbers and retrieving the corresponding titles from the CrossRef database. The following example code illustrates this test:
import time
issn_list = ["194057-58", "1940.5758" , " 19405758"]
headers = { 'User-Agent': 'ISSN-info/1.0 (mailto:myemail@email.com)'}
for issn in issn_list:
cleaned_issn = issn.replace("-", "").replace(" ", "").strip()
time.sleep(2)
crossref_url = f"https://api.crossref.org/journals/{cleaned_issn}"
crossref_response = requests.get(crossref_url, headers = headers)
crossref_data = crossref_response.json()
if 'message'in crossref_data:
journal_info = crossref_data['message']
print("Journal Title:", journal_info.get('title'))
else:
print("Not available")
As a result, regardless of the different representations of the ISSN numbers in the list, we obtained the same title information, “code4lib,” indicating the successful iteration and retrieval of corresponding titles from the CrossRef database using the for loop (Figure 9).
Journal Title: code4lib Journal Title: code4lib Journal Title: code4lib
Figure 9. Code Execution Result – Processed Titles by Iterating with a for Loop
Building upon this foundation, we extended the functionality to incorporate data requiring ISBN numbers. By implementing an if statement, we systematically assessed each item in the “issn_list” to determine whether it represented an ISSN or an ISBN. [15] If the value corresponded to an eight-digit ISSN, the code executed the necessary steps to request title information from the CrossRef database. Conversely, if the value represented a ten-digit or thirteen-digit ISBN, the code performed the relevant tasks to retrieve the title from the Google Books database. The final version of the code we deployed is presented below: [16]
""" ISSN LOOK UP"""
issn_list = list(article["ISSN"])
transaction_list = list(article['Transaction Number'])
# Initialize a new dataframe
new_df = pd.DataFrame(columns=['Transaction Number', 'issn', 'title', 'original'])
# Update the Transaction Number
for idx, v in enumerate(transaction_list):
new_df.loc[idx, 'Transaction Number'] = v
# Iterate through the transaction list and search for corresponding ISSNs and titles from CrossRef
for idx, x in enumerate(issn_list):
x = str(x)
new_df.loc[idx, 'original'] = x
x_short = x.replace("-", "").replace(" ", "").strip()
if len(x_short) == 8:
x_issn = x_short[0:4] + "-" + x_short[4:8]
api_url = f"https://api.crossref.org/journals/{x_issn}"
try:
headers = {'User-Agent': 'ISSN-info/1.0 (mailto:myexample@email.com)'}
response = requests.get(api_url, headers=headers)
data = response.json()
# Retrieve the ISSN and title from the response
if "message" in data:
journal_info = data["message"]
if "ISSN" in journal_info:
new_df.loc[idx, "issn"] = journal_info["ISSN"][0]
if "title" in journal_info:
new_df.loc[idx, "title"] = journal_info["title"]
except:
pass
# Iterate through the transaction list and search for corresponding ISBNs and titles from Google Books
elif len(x_short) == 13 or len(x_short) == 10:
try:
api_key = "-------------" # Your key here.
url = "https://www.googleapis.com/books/v1/volumes?q=isbn:" + x_short + "&key=" + api_key
response = requests.get(url)
book_data = response.json()
if 'items' in book_data:
items = book_data['items']
if len(items) > 0:
volume_info = items[0]['volumeInfo']
new_df.loc[idx, 'issn'] = volume_info['industryIdentifiers'][0]['identifier']
new_df.loc[idx, 'title'] = volume_info['title']
except:
pass
In summary, this final code encompasses the following steps. Firstly, it defines the “issn_list”, which is the list of ISSN numbers to be parsed alongside the transaction numbers. Subsequently, a new DataFrame is created to store the parsed ISSN numbers and their corresponding titles. Using a combination of a for loop and if/elif statements, the code evaluates each ISSN number to determine whether it represents an ISSN or an ISBN. Based on the identification, the code retrieves the title information from either the CrossRef or Google Books database. The resulting titles and their corresponding ISSN numbers are then stored in the newly created DataFrame.
3. Results and Discussion
By implementing our customized Python code, we achieved a substantial improvement in the accuracy of our data project. To showcase this improvement, we conducted a comparison between the original dataset and the processed dataset, specifically focusing on the top 10 most requested journal titles. The comparison revealed both minor and significant differences in the counts and specific journals between the two datasets. The table below presents the top 10 records.
| Original Data | ||
|---|---|---|
| Order | Title | Num of Requests |
| 1 | Biochimica et biophysica acta. International journal of biochemistry and biophysics 0006-3002 | 20 |
| 2 | The Journal of speech and hearing disorders : JSHD | 15 |
| 3 | Clinical pharmacy | 14 |
| 4 | Archives internationales de pharmacodynamie et de therapie | 9 |
| 5 | American journal of hospital pharmacy | 8 |
| 6 | Journal of the American Dietetic Association | 8 |
| 7 | Psychosomatic medicine | 8 |
| 8 | The Journal of laboratory and clinical medicine | 8 |
| 9 | Journal of reproductive medicine | 7 |
| 10 | Zhurnal obshcheĭ khimii | 7 |
| Processed Data | ||
| Order | Title | Num of Requests |
| 1 | Biochimica et Biophysica Acta | 22 |
| 2 | Journal of Speech and Hearing Disorders | 15 |
| 3 | Clinical pharmacy | 14 |
| 4 | Gestalt Review | 11 |
| 5 | JAMA | 10 |
| 6 | Journal of Laboratory and Clinical Medicine | 9 |
| 7 | American journal of hospital pharmacy | 9 |
| 8 | Psychosomatic Medicine | 9 |
| 9 | Archives internationales de pharmacodynamie et de therapie | 9 |
| 10 | Журнал Общей Химии | 8 |
Table 1. Top 10 Journal Title Comparison (Original vs. Processed Dataset)
The top three records showed minimal variation. For instance, the most requested journal titled “Biochimica et biophysica acta. International journal of biochemistry and biophysics 0006-3002” had a count of 20 requests in the original data, whereas in the processed dataset, it appeared as “Biochimica et Biophysica Acta” with a count of 22 requests.
However, notable disparities were observed in the fourth and fifth records. In the processed data, “Gestalt Review” emerged as the fourth most requested journal with 14 requests, whereas it did not appear in the top requested list of the original dataset. Upon investigating the original records using the Transaction Number, we discovered that variations in how users entered the journal name resulted in it being listed as several different journals in the data (Figure 10). Examples of these variations include entries with different capitalizations, variations in subtitle abbreviation, and the presence of punctuation.
58 Gestalt Review 61 Gestalt review : a publication of the Gestalt International Study Center. 323 GESTALT REVIEW. 324 Gestalt review. 697 Gestalt Review 698 Gestalt Review 699 Gestalt Review 701 Gestalt Review 776 Gestalt Review 990 Gestalt Review 1136 Gestalt review : a publ. of the Gestalt International Study Center.
Figure 10. Variations in Title Entries for "Gestalt Review"
Similarly, while “JAMA” did not appear in the top 10 list of the original dataset; it ranked fifth in the processed dataset with 10 requests. Upon examining the original records, we identified similar issues in how users entered the journal name, such as inconsistencies in the use of punctuation, and the absence of words like “the” in front of the subtitle (Figure 11).
291 JAMA : the journal of the American Medical Association. 500 JAMA : the journal of the American Medical Association. 1131 JAMA : the journal of the American Medical Association. 1248 JAMA: Journal of the American Medical Association 1456 JAMA : the journal of the American Medical Association 1497 JAMA : the journal of the American Medical Association 1523 JAMA : the journal of the American Medical Association 1547 JAMA : the journal of the American Medical Association 1593 JAMA : the journal of the American Medical Association 1653 JAMA : the journal of the American Medical Association
Figure 11. Variations in Title Entries for “JAMA”
Additionally, when considering the dataset for April 2020 alone, we found that 992 out of 1629 records had modified titles after applying our customized code (Figure 12). This corresponds to approximately 60% of the titles being changed through our data cleaning and standardization process. The number of unique journal titles decreased from 1,151 to 1,097, as well. These findings demonstrate the effectiveness of our data processing approach in cleaning and standardizing journal title data.
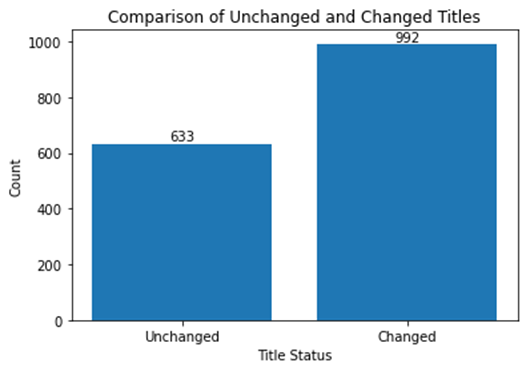
Figure 12. Bar graph Showing the Impact of Data Cleaning
Once the data is clean and standardized, the value of the data increases, enabling the data’s use in various applications. In our case, we harnessed the power of the clean and processed data to integrate it into the PowerBI visualization tool, facilitating streamlined data visualization and enabling prompt identification of patterns and trends (Figure 13). Notably, we observed significant fluctuations in the request volume for certain journals over time. For instance, there was a substantial increase in requests for medical journals like The Lancet during the COVID-19 outbreak in 2020, followed by subsequent decline in request volume. These insights provided a comprehensive understanding of journal request dynamics and facilitated data-driven decision-making [17].
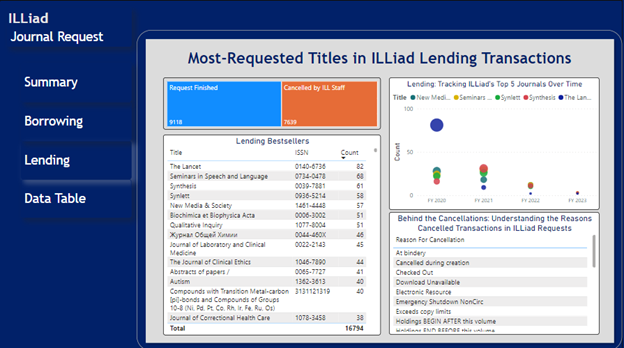
Figure 13. ILLiad Journal Request Dashboard. Notably, the size of the blue bubble is larger compared to other bubbles on the dashboard in FY2020. Additionally, the size of this blue bubble appears to decrease in consecutive years, indicating a trend of decreasing journal requests for “The Lancet” over time.
In terms of maintaining this dashboard, our customized code for data cleaning and standardization ensures consistent data formatting and sustainability, eliminating the need for additional manual labor for regular updates.
While our data processing approach has achieved significant accomplishments and improvements, it is important to acknowledge certain limitations and consider future enhancements. Although our Python code has effectively cleaned and standardized the data, there is still potential for further modifications to enhance its annotation capabilities and facilitate collaborative work. Additionally, the development of Graphical User Interface (GUI) applications can provide added convenience and accessibility. Considering the constraints of time and resources, we have prioritized simplicity and efficiency in our current code implementation, which has proven valuable for our workflow. However, moving forward, we will explore opportunities for code optimization and the development of user-friendly interfaces to enhance the efficiency and accessibility of our data processing and analysis tasks.
4. Conclusions
This study demonstrated the importance of clean and standardized data in library data projects. Clean and standardized data is invaluable in any data analysis project, including library data projects. It not only lays the groundwork for analysis but also enhances the quality of insights and facilitates more effective data utilization. By implementing the customized python code, we successfully processed and cleaned journal title data, resulting in improved accuracy and efficiency. The combination of string manipulation techniques, API requests, and automation through for loops allowed us to handle ISSN and ISBN variations effectively and retrieve the corresponding titles. Future enhancements should focus on maximizing code annotation capabilities and developing user-friendly interfaces to further enhance efficiency and collaboration.
References
ACRL. 2006. Standards for Libraries in Higher Education. Association of College & Research Libraries (ACRL). [accessed 2023 Jun 26]. https://www.ala.org/acrl/standards/standardslibraries.
Atlas Systems. 2019. Reports and Statistics. Atlas Systems. [accessed 2023 Jun 26]. https://support.atlas-sys.com/hc/en-us/articles/360011809534-Reports-and-Statistics.
Batalha F. 2023. A Python Library that Implements the Crossref API. [accessed 2023 Jun 26]. https://github.com/fabiobatalha/crossrefapi.
Chamberlain S. 2023. Client for Crossref Search API. [accessed 2023 Jun 26]. https://github.com/sckott/habanero.
Crossref Python commons library. 2022 Aug 8. GitLab. [accessed 2023 Jun 26]. https://gitlab.com/crossref/crossref_commons_py.
Crossref Unified Resource API 0.1. [accessed 2023 Jun 26]. https://api.crossref.org/swagger-ui/index.html.
imyem7. 2023. duq_open. [accessed 2023 Jun 26]. https://github.com/imyem7/duq_open.
JSON Encoder and Decoder. Python documentation. [accessed 2023 Jun 26]. https://docs.python.org/3/library/json.html.
Moon-Chung JY, Behary R, Slimon L. LibGuides: Gumberg Library Analytics: Gumberg Library. [accessed 2023 Jun 26]. https://guides.library.duq.edu/dashboards.
Overview | Google Books APIs. Google for Developers. [accessed 2023 Jun 26]. https://developers.google.com/books/docs/overview.
Pandas – Python Data Analysis Library. [accessed 2023 Jun 26]. https://pandas.pydata.org/.
pandas.read_csv — pandas 2.0.3 documentation. [accessed 2023 Jul 16]. https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.read_csv.html.
Pandas.read_excel — pandas 2.0.2 documentation. [accessed 2023 Jun 26]. https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.read_excel.html.
Petrick J. 2002. Electronic resources and acquisitions budgets: SUNY statistics, 1994‐2000. Collection Building. 21(3):123–133. doi.org/10.1108/01604950210434560.
Python Documentation: Compound Statements. Python documentation. [accessed 2023 Jun 26]. https://docs.python.org/3/reference/compound_stmts.html.
Regular expression operations. Python documentation. [accessed 2023 Jun 26]. https://docs.python.org/3/library/re.html.
Reitz K. requests: Python HTTP for Humans. [accessed 2023 Jun 26]. https://requests.readthedocs.io.
Notes
[1] “ILLiad Activity” dashboard, which is available at https://guides.library.duq.edu/dashboards, is updated monthly and contains data from January 2017 to the present. (Moon-Chung et al.).
[2] Pandas is a fast, powerful, and user-friendly open-source data analysis and manipulation tool for Python. Pandas documentation is available at https://pandas.pydata.org/docs/ (Pandas – Python Data Analysis Library).
[3] The requests library is a widely-used Python module for making HTTP requests and interacting with web APIs. The documentation is available at https://pypi.org/project/requests/ (Reitz).
[4] The json library is a built-in module in Python and it allows for easy conversation between JSON strings and Python objects. The official Python documentation is available at https://docs.python.org/3/library/json.html (JSON Encoder and Decoder).
[5] Box refers to a cloud-based file sharing and collaboration platform. https://www.box.com/ .
[6] For further information and details, refer to the documentation available at https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.read_csv.html (pandas.read_csv — pandas 2.0.3 documentation)
[7] The pandas.read_excel function offers support for reading file extensions such as xls, xlsx, xlsm, xlsb, odf, ods, and odt from both local filesystems and URLs. For further information and details, refer to the documentation available at https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.read_excel.html. (Pandas.read_excel — pandas 2.0.2 documentation).
[8] The complete code with sample data is available on the project’s GitHub page, https://imyem7.github.io/duq_open/ (imyem7 2023)
[9] The re module in Python enables pattern matching, searching, and manipulation of strings in a flexible and efficient manner using regular expressions. Documentation is available at https://docs.python.org/3/library/re.html (Regular expression operations)
[10] Please make sure to refer to the full Crossref REST API documentation for detailed instructions on its usage. The documentation is available at https://api.crossref.org/swagger-ui/index.html (Crossref Unified Resource API 0.1).
[11] The documentation for crossref-commons is available at https://gitlab.com/crossref/crossref_commons_py (Crossref Python commons library 2022 Aug 8).
[12] Habanero also offers features such as citation counts and DOI resolution. The documentation is available at https://github.com/sckott/habanero (Chamberlain 2023).
[13] The documentation for crossrefapi is available at https://github.com/fabiobatalha/crossrefapi (Batalha 2023).
[14] For further information about the Google Books API and its capabilities, please visit their official documentation at https://developers.google.com/books/docs/overview (Overview | Google Books APIs).
[15] If you would like to learn more about compound statements in Python, please visit https://docs.python.org/3/reference/compound_stmts.html for further information (Python Documentation: Compound Statements).
[16] The complete code with sample data is available on the project’s GitHub page, https://imyem7.github.io/duq_open/ (imyem7 2023)
[17] The ILLiad Journal Request dashboard is available at the Gumberg Library Analytics page, https://guides.library.duq.edu/dashboards (Moon-Chung et al.)
About the author
Jennifer Ye Moon-Chung is a digital projects and analytics manager at Duquesne University, and an MLIS candidate focusing on Applied Data Driven Methods (ADDM) at the University of Pittsburgh.


Subscribe to comments: For this article | For all articles
Leave a Reply