By Adrienne VandenBosch (0000-0001-8777-8699), Keith E. Maull (0000-0002-3459-5810), and Matthew Mayernik (0000-0002-4122-0910)
Introduction
Jupyter Notebooks have seen widespread adoption in scientific disciplines, from geosciences and astrophysics, to computational biology and others. Their ease of use, support for the Python programming language (among others), portability, shareability and easy deployment onto a wide range of hardware and into execution environments from local to remote, has made them popular among novice and expert users alike. The proliferation of notebooks, however, has produced a number of challenges. The enthusiasm for sharing notebooks on software repository platforms like Github (https://github.com), have made things increasingly complex. Github, while a platform likely to exist for a long time, is not an institutional repository (IR), and does not share the same commitment to preservation and permanence as traditional IRs. To address this issue, the Software Heritage service (https://www.softwareheritage.org/) has been created to preserve the contents of Github repositories, among others. It operates on a massive scale, duplicating Github at large without any filtering. It thus provides preservation capabilities, but not access, curation, or support. Therefore, we ask an important question for scholarly libraries: how do institutional repositories play a role in preserving these important digital outputs of scholarly work and making them more accessible and usable? Institutional repositories typically house scientific and scholarly output as the permanent record of comprehensive scholarship amongst other related and relevant digital assets such as publications or data. It is therefore not unreasonable for traditional IR platforms to play an active role in supporting notebooks despite their relative novelty in the modern scholarly workflow. The challenges of doing so are significant, but the tools, techniques and technologies already exist for doing so.
Thus, in this paper we investigate what it might take for notebooks to be accessioned into institutional repositories as first-class scholarly assets. Many questions remain around just how these new outputs of 21st century scholarship fit into the landscape of other more traditional assets like publications or even data. It is clear, however, that before we can understand their place among assets which are already accepted, we must address one of the primary barriers to accepting notebooks into traditional repository infrastructure: metadata. Whether inconsistent or altogether absent, any digital asset without proper and appropriate metadata is as good as a ghost within the wide and vast repositories operated by the best academic institutions around the world.
The optimal solutions for managing, preserving, and providing access to computational notebooks lie within some combination of tools, standards, best practices, guidelines, incentives, penalties or any number of remedial strategies to help certain communities realize the importance of these digital products. But which tools and standards? What best practices? Whose guidelines? At the moment, there are standards and best practices that could be applied, yet little consensus has grown around their adoption and use. Likewise, tools and schemas that could generate appropriate metadata are either not widely used or known to end users. Furthermore, many standards, guidelines, and best practices require that communities adopt them, and often this is driven by governance structures and community consensus.
More needs to be done to accelerate and extend the range of existing solutions, best practices and tools, as well as consider more rigorous approaches to provide the highest value for notebooks to take their place among more traditional digital assets within institutional repositories. In this paper, we will:
- detail the state of Jupyter notebooks (and more generally, executable code) in the context of managed institutional repository assets through careful examination of best practices and community knowledge,
- explore how existing institutional repositories are implementing the accessioning of notebook assets,
- examine the concrete application of CodeMeta schema metadata mappings to Jupyter notebooks, and
- explore how a single scientific institution, the National Center for Atmospheric Research (NCAR), might assess and prepare a collection of their own Github notebooks for accession and permanent deposit into their OpenSky Institutional Repository.
Given the relative sparseness of work on Jupyter Notebooks in IRs, this project is necessarily exploratory of a broad landscape. But we hope that our work can point out current trends, challenges, and opportunities that offer insight for next steps within the digital library community.
Jupyter Notebooks
Jupyter notebooks have become the de facto tool in many modern data science workflows, and have become an important tool in education and training across many disciplines. Their popularity is owed to the confluence of several factors. First, the rise of computational thinking in modern scientific disciplines is now unavoidable (Wing, 2011). The size and scope of data requirements to conduct scientific inquiry require active application of computational techniques. While the use of computation in science is not new, the scope of its use is, requiring more domain experts than ever to learn how to write software to conduct the kind of research necessary to make field-relevant advances. The Python programming language is the second factor contributing to the broad adoption of Jupyter notebooks. Python is a language that was initially developed as an alternative to languages like C to lower the syntactic complexity and simplify the semantic expressiveness of programs. While over 30 years old as of 2023, in the past decade, Python has enjoyed a dramatic increase in popularity and use among computational newcomers because of its simple and unimposing syntax, natural expressiveness, intuitive data structures and robust software libraries for a broad range of scientific disciplines from astrophysics, genomics, geoscience to others.
Jupyter notebooks are executed within a portable, web-based platform that combines into a single tool the execution capabilities of typical programming interfaces, with the expressive capabilities of narrative structure. A notebook consists of a series of cells that contain program fragments intended to be executed sequentially. The program fragments can contain any valid code. Though Jupyter supports many more languages, Python is the most popular within the platform. A cell may also contain non-code fragments of text, possibly technical expositions that either expand details of the code, or provide theoretical or any other explanation of the contents of the notebook. When seen as a whole, these cells form the “notebook”, and a form of what can be called computational narrative.
Notebooks have a tremendous amount of appeal because, not only do they capture the execution of the cells themselves (and hence the “executable” nature of notebooks), but they may contain a variety of cell outputs such as images, diagrams, graphics and even interactive widgets that allow the end user to actively change inputs and see corresponding changes in real-time (see Figure 1 for an example). Many feel that these notebooks and their interactive features hint at the future of all scientific explanation that provides the transparency and auditability necessary for computational-centric science to progress (Gil et al., 2007; Stodden et al., 2016; Stodden et al., 2018; Brinckman et al., 2019). Indeed, on platforms like Github, there are now millions of such notebooks (Rule et al., 2018), some of which contain important scientific exposition. Jupyter notebooks of scientific value are the ones that we are most interested in for this work, and specifically, how they become part of the permanent scientific record by way of IRs.
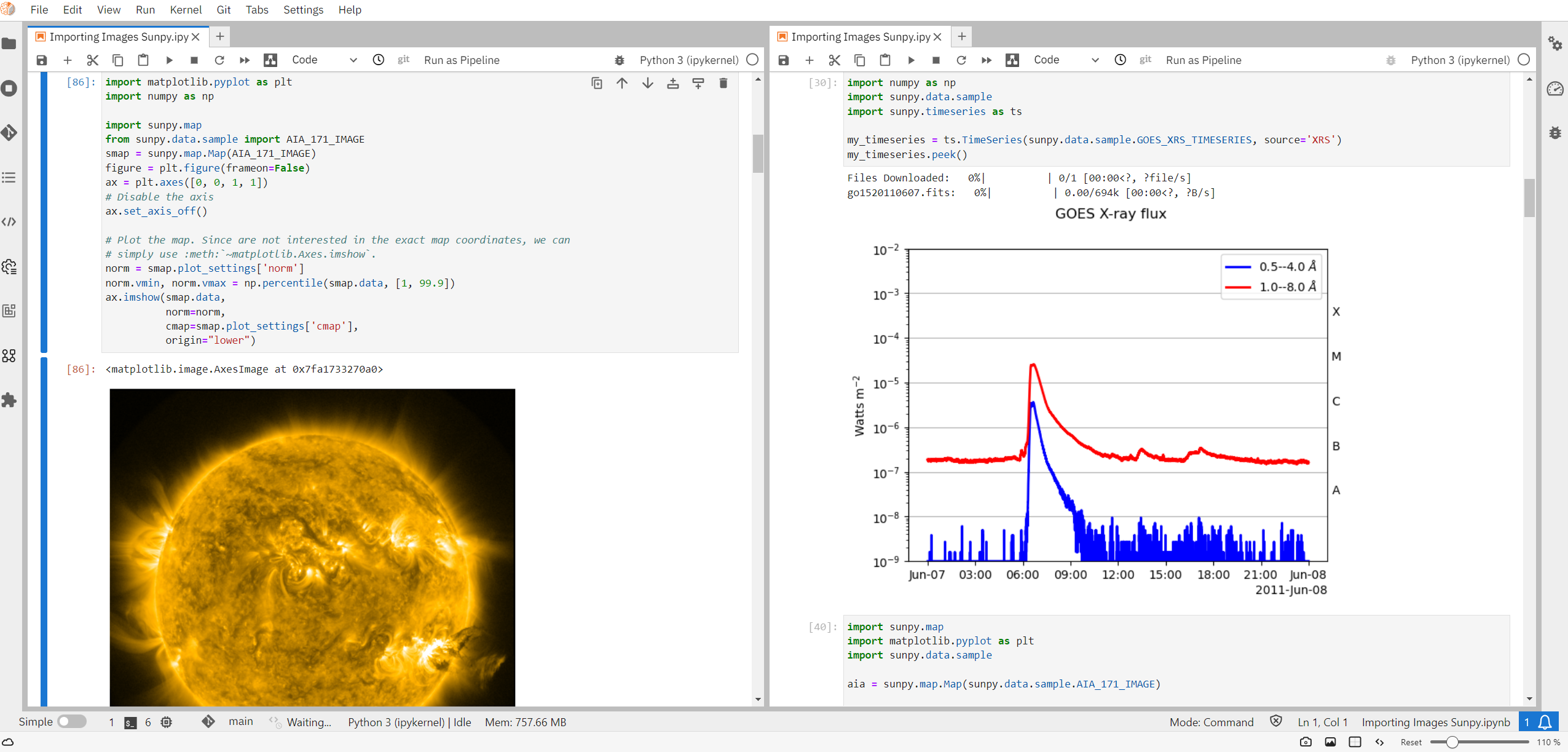
Figure 1. Example Jupyter Notebook.
Jupyter Notebooks and Institutional Repositories
Jupyter Notebooks are exploding in number and variety, and are being used as part of scientific workflows and in other data science applications. These resources are highly variable in size, content, and utility, however (Wang et al., 2020). Many Jupyter Notebooks that are publicly available on Github appear to serve as exploratory tools for their creators, and have not been created with any intention of use by anybody but the creator (Rule et al., 2018). Further, despite their promise as sharable resources that can be reused and re-executed by known or unknown collaborators, many publicly available Jupyter Notebooks have problems that prevent them from being re-executed in any straightforward fashion (Pimentel et al., 2021). For example, many notebooks do not indicate the versions of the libraries used, making precise reproduction tricky. Additionally, when there are errors due to missing files, for example when data files are imported using hard-coded paths instead of relative or URL paths, re-execution, let alone reproduction becomes even more challenging.
Institutional repositories come into the discussion as a potential home for long-term curation and preservation of Jupyter Notebooks. This discussion is stimulated by the widespread use of Github as a place to store and share Jupyter Notebooks, and by the increase in links to Github within scholarly publications (Escamilla et al., 2022). Github has many great features to support software development and file sharing generally. But it is owned by Microsoft, a for-profit corporation, and is not designed or operated to be a preservation environment. Microsoft has made gestures toward preservation of material hosted in Github, such as creating an “Arctic Code Vault,” in which a snapshot copy of Github was deposited into a frozen Norwegian mine in 2020. But such publicity stunts belie the lack of any practical digital preservation strategies built-in to Github (Rosenthal, 2019).
Can IRs have a role in addressing this challenge? Although the IR landscape has evolved significantly over the past 20 years, the basic functions of IRs remain consistent with Clifford Lynch’s formulation from 2003 (pg. 328): an IR consists of “a set of services that a university offers to the members of its community for the management and dissemination of digital materials created by the institution and its community members. It is most essentially an organizational commitment to the stewardship of these digital materials, including long-term preservation where appropriate, as well as organization and access or distribution” (Lynch, 2003). This organizational commitment has thus far focused on publications, gray literature, and digital data sets. Where do software and computational notebooks fit within organizational commitments regarding digital curation? It may be that IRs are not the optimal approach for curation and preservation of Jupyter Notebooks. Perhaps centralized services like Zenodo (https://zenodo.org/) and Software Heritage have benefits regarding economies of scale and sustainability planning, but this is an open point of debate (Arlitsch and Grant, 2018). One central finding of IR research and operations, however, is that IRs are continuously needing to be reassessed, iterated, and evolved over time, with this evolution encompassing the IR technologies, scale and scope, services, and the collections themselves (Cragin et al., 2010; Borgman et al., 2015; Fallaw et al., 2021).
In the remainder of this paper, we will survey the terrain of software for institutional repositories along four strata. Stratum one explores the community knowledge and best practices within IR communities for software assets. Given the increased general demand for software deposits, a unique set of community behaviors and knowledge has emerged that requires deeper understanding. Along stratum two we narrow the scope of investigation to five large IRs to learn more about what concrete repository practices can be observed for Jupyter notebook deposits in the wild. Learning how best practices are synchronized with implementation provides important links to the realities facing IRs. Within stratum three we dive deeper into CodeMeta (https://codemeta.github.io/) as an important tool for capturing Jupyter notebook metadata. Here we explore how this schema can be crosswalked to the Metadata Object Description Schema (MODS). This effort explores how software-focused metadata might fit within typical IR metadata structures. In the fourth stratum, we examine how a single IR might act as a case study in decision making by looking at the National Center for Atmospheric Research (NCAR) Github organization for Jupyter notebook candidates that might be deposited into the IR at NCAR. This exercise demonstrates some of the complexity of the decision to accession notebooks, as well as the nuance required to be successful.
Community Knowledge and Best Practices
An investigation of community knowledge was conducted through the analysis of relevant literature to gather current ideas, best practices, and theories about software and executable code as assets in IRs. Publications regarding the use of Jupyter Notebooks as scientific objects were also analyzed to gather information on how creation and use may affect decisions around managing them in IRs.
Jupyter Notebooks are used for analysis of results, as supplements for publications and as the source of books, dissertations, theses and in many other research contexts (Kluyver et al., 2016). Increasingly, they are being integrated into and may form the backbone of complex scientific workflows (Sullivan et al., 2019) and hence the target of recommendations for how such workflows are reproduced and what tools to use.
A typical notebook is run in a specific computational environment with an underlying hardware and software configuration. Specific versions of modules and libraries that are necessary for proper (and expected) functioning of a notebook are crucial, since if even versions of imported modules are different, execution outputs, and hence reproducibility and importantly replicability may be impossible, leading to undesirable outcomes that are increasingly plaguing scientific research (Reality check on reproducibility, 2016; Resnik and Shamoo, 2017). Capturing, codifying and standardizing these configurations has been the topic of much research in software generally and Jupyter notebooks, specifically. For example, Bouquin, et al (Bouquin et al., 2019), suggest that notebook metadata include version information for Jupyter kernel and distribution, while Erdmann, et al (Erdmann et al., 2021) recommend, in addition to module versions and dependencies, that zero-install cloud environments and configuration be considered to facilitate running and executing notebooks reliably. Still too, Beg, et al (Beg et al., 2021) recommend using MyBinder (https://mybinder.org/) to ensure notebooks are rerunnable, but acknowledge that relying on third party tools may introduce unresolved complications. Similarly, containerization methodologies such as Data and Software Preservation for Open Science (DASPOS) (Thain et al., 2015) provide a glimpse of what might be considered (and which tools to use) to specify, assemble and execute software, including all dependencies. These are just a few of the ongoing efforts to address the executable reproducibility of notebooks.
Storing notebooks and their corresponding metadata is necessary for discoverability and preservation. At the moment, many Jupyter notebooks are created (and hence stored) on the Github platform, which provides reliable, yet impermanent storage for notebooks. Notebooks stored on Github can be removed at any time by their creator (and infrequently, by Github), and while the platform provides an incredible level of detail about changes to a repository and any other asset in them, an internal Github-maintained metadata schema and data format for storing and tracking data is used.
Concerns about longevity and metadata of assets in Github have encouraged several community efforts. For example, Zenodo is a platform supported by the European Commission (EC) and operated by OpenAIRE and CERN (Conseil Européen pour la Recherche Nucléaire) that focuses on permanence and preservation of Github repositories. Not only does the Zenodo platform provide automated mechanisms for providing versioned snapshot archives of Github repositories, it provides persistent identifiers (as DOIs) and metadata in schemas common to institutional repositories, such as MARCXML, Dublin Core (DC) and DataCite. When used in conjunction with recommendations like those suggested by Greenberg, et al (Greenberg et al., 2021), Zenodo reveals an important step that shows how normalizing both end user deposit behavior, best practices and the structure of metadata might inform institutional repository decision making for software as well as notebooks.
Unified frameworks for software citation and attribution have been proposed by FORCE11 (Katz et al., 2019) who identify six essential dimensions of software citation: (1) importance, (2) credit and attribution, (3) unique identification, (4) persistence, (5) accessibility, and (6) specificity. These same dimensions are applicable to Jupyter notebooks, and provide guidelines for curation, standardization of metadata and deposit of notebooks along these dimensions. For example, given the broad proliferation of notebooks, importance criteria are especially necessary for institutional repository curators to scrutinize what is deposited, as resource limitations require adequate filters on accessioned assets. Furthermore, credit and attribution as well as unique identification have been the subjects of robust debate. DOIs (Digital Object Identifiers) have been used since their creation as a robust mechanism to ensure the attribution, persistence and resolution of digital objects, and are recommended as the preferred identifier for many assets deposited into IRs (Banerjee and Forero, 2017), though other persistent identifiers (e.g. handles) could be equally valid in many IR contexts. However, DOIs typically resolve to a single digital object that is not subject to significant change (e.g. a publication), yet software in general is often changing and the versions of those changes are important for research, putting unique pressure on the concept of using a single DOI for an evolving object. Recommendations for citing versions of software often follow patterns established for versioned data citation, though consensus on the accepted strategy for minting DOIs for software versions has yet to emerge. Furthermore, unique challenges emerge with the use of microcitation or highly granularized citations that might point to individual lines of software source code (data subsets in the case of data) or in the case of Jupyter notebooks, output cells containing figures, graphs or computation outcomes. It is acknowledged that this type of citation is more difficult to achieve with a DOI, and in many research communities where dataset (subset and derivative) citation is acknowledged as necessary for research reproducibility (such as Earth and life sciences communities), important recommendations (Patrinos et al., 2012; ESIP Software Guidelines Draft, 2016) have emerged to address some of the underlying issues. For software, these issues have been aggressively addressed by the non-profit organization Software Heritage to focus on the unique characteristics of software and the corresponding wide range of citation use cases. Their SWH-ID URI schema (Di Cosmo, 2020), which also permits microcitation, is an important step forward, even if it breaks from the conventional use of DOIs.
Important work in deposit guidelines for software digital assets has been innovated by the Software Sustainability Institute (Brown et al., 2018), who provide a detailed framework for why, when, what, where and how software might be deposited into a repository by researchers and curators alike. Their framework proposes that a deposit adhere to one of three levels: minimal, runnable and comprehensive – where a comprehensive deposit would not only include the most detailed metadata, but also detailed documentation about the software, its purpose, function, and ecosystem (e.g. dependencies) as well as test code such that expected outputs are well understood and inspected against running code. This can be especially important in a Jupyter notebook context, where a notebook can be deposited with the executed cells as well as unexecuted cells, and if a notebook is executed and any cells fail, it can be considered as potentially unfit for deposit according to SSI guidelines. Knowing whether a notebook has failing cells would also be necessary to develop a larger strategy for assessing quality standards of deposits.
Similar efforts to encourage software deposit and lift software to first class status can be seen with the peer-reviewed Journal of Open Source Software (JOSS) (Smith et al., 2018), which accepts software submissions of all kinds to normalize software as an important asset in scientific communication at the same level of importance as publications. The journal enforces systematic and consistent structure, description and execution standards of software submissions to encourage a community of practice built around high quality software norms and guidelines. While deposits into JOSS stay within their transparent community-centered platform, any of its deposits could be accessioned into a relevant IR, but importantly, JOSS demonstrates a comprehensive approach to deposit, metadata, preservation and presentation – which might inform IRs of strategies that may be implemented in their own environments adapted to the needs of their specific communities.
Finally, metadata for notebooks is an important, but often neglected topic. Building more on the Software Sustainability Institute’s typology of minimal, runnable, and comprehensive software deposits, metadata for software minimally requires README files with the software name, description, contact information, and link to the live project or source code; they should also include explicit copyright and license information, author and contributor information, and the source code (Jackson, 2018). For a runnable deposit, a PDF or static HTML version of the notebook, a full dependencies list, a citation file, a CodeMeta/JSON-LD file and a container file where applicable (Rule et al., 2018 Oct). In a runnable deposit the README should include a version number, copyright statement, user documentation, and a summary of other deposit content (Jackson, 2018). Even without runnable or executable software, source code can have long-term value as data (ESIP Software Guidelines). Runnable and comprehensive deposits include richer metadata about dependencies and programming languages, more in depth documentation, sample data, and a containerized version/emulation of the software. One set of metadata guidelines explicitly focused on software is the CodeMeta framework. We discuss CodeMeta in detail in a later section, as it defines metadata elements that are most relevant, adequate and appropriate for software.
All of these efforts reveal the inherent complexity and limitations of current strategies for citing, describing and preserving software, and more generally the unique features of emerging digital objects that must be curated and preserved as part of the 21st century scientific record. While many of these efforts are maturing, and their communities still growing, there exists substantial demand and enthusiasm to build techniques, tools and strategies that demonstrate the importance of software and executable digital objects such as Jupyter notebooks.
Jupyter in Existing IRs
We looked at the institutional repositories of five large research institutions to see what examples of Jupyter Notebooks in IRs already existed and what could be learned from them. Caltech Data, Michigan Deep Blue Data, Stanford SearchWorks, Virginia Tech Data Repository, and University of California San Diego Digital Collections were chosen because they were known to have software or notebook files. This was not intended to be a systematic examination of the entire IR landscape, but to provide insight into key characteristics of Jupyter Notebooks as they exist in IRs. From these observations, we found that deposits containing Jupyter Notebook files did not follow uniform practices, even when existing in the same IR. In one repository, Notebooks were categorized as three different types of objects (data, software, and workflow), perhaps referencing their intended use case and therefore a need for different preservation practices. Trying to understand decisions made regarding the handling of notebooks and related objects was especially challenging since the IRs had few notebooks to compare for consistency of practices. Future investigation would likely benefit from additional insight into guidelines and decisions for preservation for these items.
Table 1 summarizes the characteristics of the Jupyter Notebooks that existed in the IRs we examined. A few deposits included a static representation of the executed notebook. In the majority of cases this was as a PDF, although one included a text file of the notebook JSON. This follows recommendations for including static representations, so that even if the notebook is no longer executable, there is some sort of representation of its original content. Other IR deposits in the results of our search were data sets, mentioning ipynb or Jupyter Notebooks in their description, and linking to GitHub repositories where the actual notebook files were stored and could be previewed. The Virginia Tech IR was powered by Figshare (https://figshare.com/), which allows for an in-browser preview of ipynb files. Here, a search for the python language unearthed a result that contained 6 zipped ipynb files that did not show up in searches for ipynb or jupyter. One repository, Caltech Data, provided links to MyBinder for resources that contain an ipynb file, which enables users to invoke an online environment that allows for viewing, editing, and running the notebook file (Morrell, 2019). Caltech Data has since moved to a new platform where myBinder links do not currently appear to be supported.
*Limited to the resource type of ‘dataset’, due to full integration of the library catalog.
**No individual ipynb files were available to test interactions, consisting of compressed/non-ipynb files.
| IR | # results for ipynb | # results for Jupyter | # results for python | Interaction, Method | Metadata |
| Caltech Data | 9 | 14 | – | Invocable, Binder | Not discernible |
| Stanford SearchWorks* | 2 | 2 | 19 | Preview, catalog** | MODS |
| Virginia Tech | 1 | 1 | 2 | Preview via Figshare | DataCite, Dublin Core, NLM (figshare options) |
| Michigan Deep Blue Data | 1 | 2 | 19 | Preview, catalog** | Not discernable |
| UC San Diego | 4 | 32 | – | Preview, catalog** | Not discernable |
Metadata schemas were unable to be determined for the majority of the repositories that we examined, making it difficult to find out if the information needed to access or support reproducible notebooks would be available. With most of the files in compressed form, searching for .ipynb would not necessarily return all results that contain Jupyter Notebooks without a complete list of files in a field that is searchable in the online catalog. Once located, there is still the question of whether or not these notebooks are executable — are they provided with the information needed to maintain their relationships to external objects? Notebooks require specific versions of external objects (libraries, data, etc.) and a particular ordering of cells to be reproducible. In the few instances where metadata schemas were distinguishable, it is clear that end users would struggle to find the necessary information to make these objects function.
CodeMeta Schema For Jupyter
What metadata should exist for Jupyter Notebooks? Examples of relevant metadata for Jupyter-based tools might include metadata about the inputs to the notebooks (data), the products or outputs of the notebook cells, the structure of the notebook itself, including linked or supplemental notebooks and, of course, metadata about the tools themselves (Leipzig et al., 2021).
Jupyter Notebooks are generally categorized as software, thus metadata recommended for software provides a good starting point in considerations for metadata that enable the preservation and reuse of these resources. The CodeMeta (Jones et al., 2017) metadata framework provides one of the most robust platforms for describing software using JSON-LD and XML. For example, it has been used in the R community to automatically generate metadata for R software packages (Boettiger, 2017). It is appropriate to discuss it regarding Jupyter notebooks, because it captures a broad range of features that target the essential concerns for notebook content, including notebook configuration, such as language versions, target operating platforms and software dependencies.
A crucial challenge remains unsolved, however: crosswalking CodeMeta to schemas commonly found in IRs (MODS, Dublin Core, etc.). Here we describe a crosswalk between CodeMeta and the Metadata Object Description Schema (MODS, https://www.loc.gov/standards/mods/), as a demonstration of the challenges and possibilities involved in applying the CodeMeta framework within a metadata schema that is commonly used in an IR. MODS was chosen because at the time of this writing MODS is the underlying metadata framework in the NCAR Library’s IR, called OpenSky (https://opensky.ucar.edu/). In order to complete the mapping, CodeMeta and MODS documentation were consulted in-depth. Sample XML files from three objects in OpenSky were used to verify the compatibility of crosswalk recommendations with actual repository records. A snippet of this mapping is shown in Table 2.
Table 2. Sample table of crosswalk from MODS to CodeMeta.
| CodeMeta | MODS |
| author | name/role/roleTerm(type?, =’code’)/[.=’aut’] |
| citation | relatedItem(otherType?, =’references’) |
| contributor | name/role/roleTerm(type?, =’code’)/[.=’ctb’] |
| copyrightHolder | accessCondition/rights.holder/name |
| copyrightYear | accessCondition/creation/year.creation |
| dateCreated | originInfo/dateCreated |
MODS has 20 top-level elements, while CodeMeta contains 67, and this fidelity mismatch requires any functional crosswalk to map top-level CodeMeta elements to attribute level metadata from MODS. MODS allows for multiple uses of the same top-level element in a record (e.g. multiple name tags for more than one author), that can then be distinguished using sub elements and attributes. For an item with two authors and one sponsor you would have three name tags, two with the roleTerm type as author (aut in MARC relator terms), and one with the roleTerm type as sponsor (spn). Where the mapping struggles the most is when these sub-elements and attributes do not yet have the controlled vocabulary to ensure consistency and accuracy of the crosswalk. For example, MODS defines the affiliation element as one that can contain address and email as well as organizational affiliations using that same subelement. However, there is no attribute of affiliation that allows for the direct mapping of email from MODS to the CodeMeta email property. Where these attributes and controlled vocabularies don’t yet align with CodeMeta properties, MODS does have the extension element. The MODS extension element creates an opportunity to preserve all unmapped CodeMeta properties within that element tag. This is not ideal, but would ensure that if a notebook were to be ingested from an environment using the CodeMeta into an IR using MODS, the needed information for notebook understanding and execution would not be lost. Alternate approaches could be to map key metadata fields to MODS as noted above, and upload a full CodeMeta with all of the relevant fields as a separate JSON file into the repository, or include the metadata directly in the notebook as a header cell.
Identification & Extraction: NCAR Github
When choosing to ingest from GitHub into an IR for long-term preservation and access, it is important to assess how ingest-ready a Jupyter notebook might be. The GitHub API provides access to internal metadata which serve as markers of notebook readiness. In order to see what markers and additional information would be discoverable, we used the GitHub API to look specifically at the NCAR Organizational GitHub. Limiting our assessment to the NCAR Organizational GitHub also gave us the ability to look at the variation of practices within a fairly controlled environment, putting into perspective just how much variation will likely exist in larger organizations. Summary characteristics of the Jupyter Notebooks found in the NCAR Github are shown in Table 3.
Table 3. Summary of Jupyter-centric repos in the NCAR Organizational GitHub.
| Repository Characteristic | Notebook Count |
| Repo was identified as containing Jupyter NB | 93 |
| Repo was identified as having majority of Jupyter NB language ≥ 60% Jupyter NB |
89 |
| Had Description | 70 |
| Indicated License | 51 |
| Project potentially stable/candidate for ingest Created before Jan. 1, 2020 |
39 |
| Project is current/candidate for consult Updated after Jan. 1, 2022 |
41 |
| Repo with higher interest level 25+ Watchers |
5 |
| Associated with a DOI DOI in README file, either assigned or to a related object |
8 |
Our API query found repositories within the NCAR GitHub that were “Jupyter-centric,” having at least 60% of their identified languages as Jupyter Notebook. It is important to note, that the languages in a repository are determined by GitHub and may change over time or differ from individual identification. From our query, we identified 89 of the 93 repos containing Jupyter Notebook language, with at least 60% Jupyter Notebook as the determined language. We gathered the count of watchers and forks, open issues, owner info, object license, and if a README or CFF file was present. These data, we felt, would be good indicators of the dimensions of importance through follower counts, credit and attribution through citation files and licensing information, unique identification and persistence through an assigned or related DOI, and persistence through creation date and last update as well. From the 93 repos, 51 had licenses, eight had DOIs present in the README file, and five had 25 or more watchers. The date of creation or last update for a repo could be used to identify long standing projects that might be at a stable state for ingest into an IR. Alternatively, a repo with a large following and frequent updates may indicate a project that would be a future candidate for ingest. Gathering this information could flag repositories to approach about possible ingest. This information could also provide targets where an IR could offer support to improve the quality of metadata so that deposits are received in a more complete state. Further work is necessary to assess the extent of notebook accessibility and executability, looking closer at content and metadata.
Discussion and Conclusions
In this final section, we discuss several types of considerations for archiving and providing access to Jupyter notebooks via institutional repositories. We discuss in turn conceptual, technical, process, and preservation considerations.
Conceptual considerations. One of the most important aspects of archiving and storing assets in a repository is the conceptual integrity of the object — that is, when it is accessed, does it lie with a range of acceptable and expected forms. Jupyter Notebooks can take on a broad array of forms, from pure programming content to pure narrative content, with a broad spectrum between. Deciding what a Jupyter notebook is precisely, poses many challenges, especially when trying to classify for accessioning into a repository. For example, existing repositories studied in this paper classify Jupyter notebooks in various ways. Some contain a large amount of data (or heavily rely on it). Some contain mostly software code (often processing data from somewhere else). Some expose particular processes or workflows for doing a computational task, while others are narrative in scope and contain little code or data. With such varied and flexible forms, it becomes challenging to classify a notebook as predominantly data, software, narrative, workflow, or some ratio of all of these. This complexity puts a special burden on deposit decision making, since each of these may look quite different because of the files that must be deposited into a repository to maintain the necessary conceptual integrity (and usefulness) of the notebook. Indeed, a deposit may include one, some or all of .ipynb, Python source files (.py), JSON, CSV, PDF files and more. Second, should Jupyter notebooks be dealt with as solitary or compound objects? Notebooks can be individual files, but are often part of larger collections of objects. They therefore can have important dependencies and interrelations with other software, data, and metadata. Repositories must plan and develop strategies for accommodating Jupyter notebooks as sets of objects, and for potentially managing relationships therein, or to external objects (such as data sets provided by external organizations).
Technical considerations. From a technical point of view, since Jupyter notebooks are executable and it is indeed this executability that make them so important in scientific reproducibility, repositories must consider whether to simply provide access to the Jupyter notebook files, or whether to enable users to interact with and execute those files via the IR platform (or through some third party platform). Tools like Binder (https://mybinder.org) exist for invoking Jupyter notebooks in interactive computational environments, but require financial and computational resources that may be hard to predict as they are dependent on the size and complexity of the Jupyter notebooks and associated data. Another important reality is that many projects that create Jupyter notebooks use Github to enable active software development, collaboration, discoverability and public exchange. IRs clearly have a different set of technical capabilities (and function) than Github, and as such provide different services. Enumerating these services, while providing bridges to the unique capabilities of Github (e.g. automation, enabling programmatic connections to IRs), is yet unexplored territory. For example, IRs may act as third party platforms to Github, providing services that programmatically ingest Github repositories for access and preservation purposes. This could complement the capabilities of other Github preservation services, such as Zenodo (https://zenodo.org) and Software Heritage (https://softwareheritage.org).
Process considerations. To establish an IR as a routine location to archive Jupyter notebooks, establishing selection criteria will be important. First, identifying what to preserve is of paramount importance. The explosive number of notebooks now being created by researchers in almost every academic field, make process improvement a vital focus for IRs. For example, self-deposit (e.g. Github-to-Zenodo) may be one option, where the creators of resources decide what and when to archive their materials, but since many IRs operate via a mediated deposit strategy, IRs would need to prioritize and develop clear selection and evaluation processes. Such selection and evaluation considerations also lead to another key question: what criteria are used to accept Jupyter notebooks for preservation? It is known that the quality and completeness of notebooks on Github vary greatly (Rule et al., 2018) (and elsewhere), so some standard must be developed by target IRs to guide a minimum acceptance criteria (or such exception guidelines otherwise provided). There is little value to preserving notebooks that are of low significance, incomplete, partially executable, or otherwise plagued with quality problems. IRs therefore must develop a process for evaluating and curating notebooks during the deposit stage.
Preservation considerations. In some sense, most of the considerations thus far have implications for preservation. But additional preservation considerations were uncovered through this project. First, and importantly, there is not yet a standardized metadata schema used to describe and preserve Jupyter notebooks. As noted above, connecting metadata conventions for notebooks to the metadata standards imposed by IRs is complicated. While the CodeMeta framework proposed here provides a useful starting point, it still requires many metadata schema implementation decisions, which further require expertise and careful consideration of current and future metadata needs. A second key preservation consideration is persistent identification — that is, how should persistent identifiers like DOIs be assigned to Jupyter notebooks given some of the conceptual considerations noted above? Consensus has not yet emerged on what a persistent identifier to a Jupyter notebook is pointing to — particularly when the notebook is part of a compound object. Do identifiers point to the compound object, the notebook or outputs within a notebook? How are complexities like notebook versions handled? Do identifiers maintain any integrity with the inaugural version identifier? These crucial questions do not have clear answers, but need to be addressed urgently, given the rapid acceptance of notebooks in scholarly contexts. Management processes to facilitate notebook capture, review and deposit, as well as technical processes and best practices that encourage predictable notebook structure and quality must be prioritized.
Tips for moving forward. Bringing Jupyter-based resources into IR collections will likely be easier to implement via progressive policies that provide iterative approaches that allow rapid response and adaptation to the needs of IR operators and depositors alike. Emphasizing the partnership between the IR and depositor will also lead to better outcomes. This study has uncovered the wide variance in existing practices. Several recommendations are now provided to guide IRs toward developing practices and policies for accepting Jupyter notebooks.
Recommendation #1: Develop a recommended file/folder structure for Github repositories housing Jupyter-based assets for deposit or accession into an IR.
As previously mentioned, many scholarly materials, especially those that are based on Jupyter notebooks, are stored in Github. The advantages of Github are clear for open source (and open science), but the platform itself provides little guidance on how Jupyter-based repositories should be structured. Furthermore, Github itself makes no guarantees on the long-term availability or accessibility of public code repositories, making their deposit and accession into IRs even more important. Providing a recommended template structure for Github repositories which would be deposited into an IR would accelerate and standardize Jupyter-based assets. If a deposit has its own structure different from the recommended template, it is important to encourage the depositor to reveal the details and logic of that structure so it might become part of the metadata of the deposit.
Recommendation #2: Leverage CodeMeta mappings to IR metadata schemas where possible, or develop an IR-specific schema that fits the needs of the target repository environment.
While CodeMeta has not become a standard for Jupyter-related metadata, at this time it is the most appropriate schema for these types of digital assets. Developing a custom schema is largely unnecessary as CodeMeta provides enough flexibility to integrate relevant domain and IR specific metadata. Connecting CodeMeta concepts to IR metadata frameworks still presents challenges, but the more CodeMeta is used, the more these issues can be ironed out.
There is not yet agreement whether some minimal metadata responsibility should be required of depositors or even how such metadata should be produced. Basic notebook metadata could be created by depositors and included as a file in the source Github repository containing a Jupyter notebook deposit. For example, the Citation File Format (CFF) (Druskat, 2021) for software citation metadata, might provide strategic hints for storing Jupyter metadata in machine-readable files which IR operators could then use to build a proper and more complete CodeMeta metadata as suggested in this paper. There are other technical solutions that are also possible, including embedding metadata directly into the Jupyter notebook with a plugin or extension that queries notebook authors for basic notebook metadata.
Recommendation #3: Develop and publish transparent deposit guidelines for Jupyter-based submissions that enumerate the minimal required metadata and structural requirements in recommendation #1 above.
Encouraging IR deposits of Jupyter notebooks becomes easy once a consistent Github repository structure (or any notebook submission outside of Github) is developed. When deposits are reviewed for official accession into the IR, if there are issues why submissions are not accepted, IR operators need to provide transparent processes for concrete feedback on how submissions can be improved for successful accessioning — relying as much on the published guidelines set forth to reduce ambiguity and frustration. Furthermore, providing pre-submission checklists and guides well in advance of submission, should go a long way to improve deposit quality and acceptance rates.
Recommendation #4: Develop policy guidelines that provide intuitive frameworks for decision making within IR management.
Choices and policies regarding the management of Jupyter notebooks must be informed by their relationships to external objects and their intended use cases. If the IR does not support execution of notebooks, for example, this decision should be communicated within IR management and expectations properly set.
This paper has presented a broad perspective on the landscape and challenges of modern scholarly products which should not go unnoticed by academic Institutional Repositories. Jupyter Notebooks provide an important potential use case for IRs, not only because of their wide adoption and increased growth, but also because they represent a class of digital object that is as of yet underserved by well-curated digital repositories. The variation of objects, metadata, and practices, even within singular institutions, points to an urgent need for action in developing policy and guidelines that their respective IR can support. There is yet a lot of work to be done in this area, but the time has come for IR operators and depositors alike to begin taking assets like Jupyter notebooks more seriously so that important academic products can be preserved for future generations.
Acknowledgements
We thank Peter Organisciak for input during early stages of this work. This work was supported by the Collaborative Analysis Liaison Librarians (CALL) project, made possible in part by the Institute of Museum and Library Services (#RE-13-19-0027-19). This material is based upon work supported by the National Center for Atmospheric Research, which is sponsored by the National Science Foundation under Cooperative Agreement No. 1852977.
References
Arlitsch K, Grant C. 2018. Why So Many Repositories? Examining the Limitations and Possibilities of the Institutional Repositories Landscape. Journal of Library Administration. 58(3):264–281. https://doi.org/10.1080/01930826.2018.1436778. [accessed 2023 Jan 27].
Banerjee, K., Forero, D. 2017. DIY DOI: Leveraging the DOI Infrastructure to Simplify Digital Preservation and Repository Management. Code{4}Lib, Issue 38. https://journal.code4lib.org/articles/12870. [accessed 2023 Oct 31]
Beg M, Taka J, Kluyver T, Konovalov A, Ragan-Kelley M, Thiéry NM, Fangohr H. 2021. Using Jupyter for Reproducible Scientific Workflows. Computing in Science & Engineering. 23(2):36–46. https://doi.org/10.1109/MCSE.2021.3052101.
Boettiger C. 2017. Generating CodeMeta Metadata for R Packages. The Journal of Open Source Software. 2(19):454. https://doi.org/10.21105/joss.00454. [accessed 2023 Sep 28].
Borgman CL, Darch PT, Sands AE, Pasquetto IV, Golshan MS, Wallis JC, Traweek S. 2015. Knowledge infrastructures in science: Data, diversity, and digital libraries. International Journal on Digital Libraries. 16(3-4):207–227. https://doi.org/10.1007/s00799-015-0157-z. [accessed 2023 Jan 27].
Bouquin D, Hou S, Benzing M, Wilson L. 2019. Jupyter Notebooks: A Primer for Data Curators. https://hdl.handle.net/11299/202815 [accessed 2022 Feb 1].
Brinckman A, Chard K, Gaffney N, Hategan M, Jones MB, Kowalik K, Kulasekaran S, Ludäscher B, Mecum BD, Nabrzyski J, et al. 2019. Computing environments for reproducibility: Capturing the “Whole Tale.” Future Generation Computer Systems. 94:854–867. https://doi.org/10.1016/j.future.2017.12.029. [accessed 2023 Sep 28].
Brown C, Hong NC, Jackson M. 2018. Software Deposit And Preservation Policy And Planning Workshop Report. Zenodo. https://doi.org/10.5281/zenodo.1304912 [accessed 2022 Feb 1].
Cragin MH, Palmer CL, Carlson JR, Witt M. 2010. Data sharing, small science and institutional repositories. Philosophical Transactions of the Royal Society A: Mathematical, Physical and Engineering Sciences. 368(1926):4023–4038. https://doi.org/10.1098/rsta.2010.0165. [accessed 2023 Jan 27].
Di Cosmo R. 2020. Archiving and Referencing Source Code with Software Heritage. In: Bigatti AM, Carette J, Davenport JH, Joswig M, Wolff T de, editors. Mathematical Software – ICMS 2020. Cham: Springer International Publishing. (Lecture Notes in Computer Science). p. 362–373.
Druskat S. 2021. Making software citation easi(er) – The Citation File Format and its integrations. In: Zenodo. https://doi.org/10.5281/zenodo.5529914 [accessed 2023 Sep 19].
Erdmann C, Stall, Shelley, Gentemann, Chelle, Holdgraf, Chris, Fernandes, Filipe P. A., Gehlen, Karsten Peters-von, Corvellec, Marianne. 2021. Guidance for AGU Authors – Jupyter Notebooks. https://doi.org/10.5281/ZENODO.4774440. [accessed 2022 Feb 1].
Escamilla E, Klein M, Cooper T, Rampin V, Weigle MC, Nelson ML. 2022. The Rise of GitHub in Scholarly Publications. In: Silvello G, Corcho O, Manghi P, Di Nunzio GM, Golub K, Ferro N, Poggi A, editors. Linking Theory and Practice of Digital Libraries. Vol. 13541. Cham: Springer International Publishing. p. 187–200. https://doi.org/10.1007/978-3-031-16802-4_15 [accessed 2023 Jun 2].
ESIP Software Guidelines Draft. 2016. https://esipfed.github.io/Software-Assessment-Guidelines/guidelines.html#h.pzfz7hkzxqus. [accessed 2022 Feb 1].
Fallaw C, Schmitt G, Luong H, Colwell J, Strutz J. 2021. Institutional Data Repository Development, a Moving Target. Code{4}Lib.(51). https://journal.code4lib.org/articles/15821.
Gil Y, Deelman E, Ellisman M, Fahringer T, Fox G, Gannon D, Goble C, Livny M, Moreau L, Myers J. 2007. Examining the Challenges of Scientific Workflows. Computer. 40(12):24–32. https://doi.org/10.1109/MC.2007.421. [accessed 2023 Sep 28].
Greenberg J, Hanson K, Verhoff D. 2021. Guidelines for Preserving New Forms of Scholarship. New York, NY: New York University. https://doi.org/10.33682/221c-b2xj [accessed 2022 Feb 1].
Jackson M. 2018. Software Deposit: What To Deposit. https://doi.org/10.5281/ZENODO.1327325. [accessed 2022 Feb 1].
Jones MB, Boettiger C, Mayes AC, Smith A, Slaughter P, Niemeyer K, Gil Y, Fenner M, Nowak K, Hahnel M, et al. 2017. CodeMeta: An exchange schema for software metadata. https://doi.org/10.5063/SCHEMA/CODEMETA-2.0. [accessed 2022 Jun 25].
Katz DS, Bouquin D, Hong NPC, Hausman J, Jones C, Chivvis D, Clark T, Crosas M, Druskat S, Fenner M, et al. 2019. Software Citation Implementation Challenges. arXiv:190508674 [cs]. [accessed 2022 Feb 1]. http://arxiv.org/abs/1905.08674.
Kluyver T, Ragan-Kelley B, Rez F, Granger B, Bussonnier M, Frederic J, Kelley K, Hamrick J, Grout J, Corlay S, et al. 2016. Jupyter Notebooks – a publishing format for reproducible computational workflows. Positioning and Power in Academic Publishing: Players, Agents and Agendas.:87–90. https://doi.org/10.3233/978-1-61499-649-1-87. [accessed 2022 Jun 24].
Leipzig J, Nüst D, Hoyt CT, Ram K, Greenberg J. 2021. The role of metadata in reproducible computational research. Patterns. 2(9):100322. https://doi.org/10.1016/j.patter.2021.100322. [accessed 2023 Feb 21].
Lynch CA. 2003. Institutional Repositories: Essential Infrastructure For Scholarship In The Digital Age. portal: Libraries and the Academy. 3(2):327–336. https://doi.org/10.1353/pla.2003.0039. [accessed 2023 Jan 27].
Morrell T. 2019. The Opportunities and Challenges of Research Data and Software for Libraries and Institutional Repositories. Against the Grain: Linking Publishers, Vendors and Librarians. 31(5):30–32. https://resolver.caltech.edu/CaltechAUTHORS:20200116-114211107.
Patrinos G, Cooper D, Mulligen E van, Gkantouna V, Tzimas G, Tatum Z, Schultes E, Roos M(Marco), Mons B. 2012. Microattribution and nanopublication as means to incentivize the placement of human genome variation data into the public domain. Human Mutation. 33(11):1503–1512. https://doi.org/10.1002/humu.22144.
Pimentel JF, Murta L, Braganholo V, Freire J. 2021. Understanding and improving the quality and reproducibility of Jupyter notebooks. Empirical Software Engineering. 26(4):65. https://doi.org/10.1007/s10664-021-09961-9. [accessed 2022 Jun 25].
Reality check on reproducibility. 2016. Nature. 533(7604):437–437. https://doi.org/10.1038/533437a. [accessed 2022 Jun 24].
Resnik DB, Shamoo AE. 2017. Reproducibility and Research Integrity. Accountability in Research. 24(2):116–123. https://doi.org/10.1080/08989621.2016.1257387. [accessed 2022 Jun 24].
Rosenthal, D. 2019. Seeds Or Code?. DSHR’s Blog, Nov. 19, 2019. https://blog.dshr.org/2019/11/seeds-or-code.html
Rule A, Birmingham A, Zuniga C, Altintas I, Huang S-C, Knight R, Moshiri N, Nguyen MH, Rosenthal SB, Pérez F, et al. 2018 Oct. Ten Simple Rules for Reproducible Research in Jupyter Notebooks. arXiv:181008055 [cs]. [accessed 2022 Feb 1]. http://arxiv.org/abs/1810.08055.
Rule A, Tabard A, Hollan JD. 2018. Exploration and Explanation in Computational Notebooks. In: Proceedings of the 2018 CHI Conference on Human Factors in Computing Systems. Montreal QC Canada: ACM. p. 1–12. https://doi.org/10.1145/3173574.3173606 [accessed 2023 Jan 27].
Smith AM, Niemeyer KE, Katz DS, Barba LA, Githinji G, Gymrek M, Huff KD, Madan CR, Mayes AC, Moerman KM, et al. 2018. Journal of Open Source Software (JOSS): Design and first-year review. PeerJ Computer Science. 4:e147. https://doi.org/10.7717/peerj-cs.147. [accessed 2022 Jun 25].
Stodden V, McNutt M, Bailey DH, Deelman E, Gil Y, Hanson B, Heroux MA, Ioannidis JPA, Taufer M. 2016. Enhancing reproducibility for computational methods. Science. 354(6317):1240–1241. https://doi.org/10.1126/science.aah6168. [accessed 2023 Sep 28].
Stodden V, Seiler J, Ma Z. 2018. An empirical analysis of journal policy effectiveness for computational reproducibility. Proceedings of the National Academy of Sciences. 115(11):2584–2589. https://doi.org/10.1073/pnas.1708290115. [accessed 2023 Sep 28].
Sullivan I, DeHaven A, Mellor D. 2019. Open and Reproducible Research on Open Science Framework. Current Protocols Essential Laboratory Techniques. 18(1):e32. https://doi.org/10.1002/cpet.32. [accessed 2022 Jun 25].
Thain D, Ivie P, Meng H. 2015. Techniques for Preserving Scientific Software Executions: Preserve the Mess or Encourage Cleanliness? Proceedings of the 12th International Conference on Digital Preservation. https://hdl.handle.net/11353/10.429560 [accessed 2022 Jun 24].
Wang J, Li L, Zeller A. 2020. Better code, better sharing: On the need of analyzing jupyter notebooks. In: Proceedings of the ACM/IEEE 42nd International Conference on Software Engineering: New Ideas and Emerging Results. Seoul South Korea: ACM. p. 53–56. https://doi.org/10.1145/3377816.3381724 [accessed 2023 Jan 27].
Wing JM. 2011. Computational thinking. 2011 IEEE Symposium on Visual Languages and Human-Centric Computing (VL/HCC).:3–3. https://doi.org/10.1109/VLHCC.2011.6070404. [accessed 2023 Sep 28].
About the Authors
Adrienne VandenBosch is the Reference Services Coordinator at the University of Colorado Boulder working to assess library services. Adrienne has a specialization in STEM liaison librarianship as a graduate from the cohort for the Collaborative Analysis Liaison Librarianship instructional project. At the University of Denver, she has also worked on projects for the Massive Texts Lab.
Keith E. Maull is a data scientist and software engineer at the National Center for Atmospheric Research in Boulder Colorado, USA, where he has held a position for 10 years in the NCAR Library exploring the scientific research impacts of the organization. His research lies at the intersection of open science, open data and open analytics and he has spent significant time exploring the value and impact of computational narratives and Jupyter on 21st century science.
Matthew Mayernik is a Project Scientist and Research Data Services Specialist in the NCAR/UCAR Library. He is also the Deputy Director of the NCAR Library. His work is focused on research and service development related to research data curation and digital scholarship broadly. He has taught research data curation courses at the University of Denver and University of Washington. He is the Joint Editor-in-Chief of the Data Science Journal.


Subscribe to comments: For this article | For all articles
Leave a Reply