by Hannes Lowagie and Julie Van Woensel
Introduction
As the National Library of Belgium (KBR), we classify every book that comes in through legal deposit. Classification and subject indexing crucial for organising our vast collection and facilitating information retrieval for our patrons. To streamline this process, we have explored various approaches, including using ANNIF [1] and the Microsoft category classification model [2]. Each approach has its strengths and weaknesses, leading us to seek a solution that offers both ease of use and maintainability.
This paper delves into our initial exploration of using Python scripts to establish a reference dataset and automate the classification process. The reference dataset script uses a CSV export of existing records that already have a manually assigned classification and generates a new CSV file with associated words per classification category. The classification script takes MARCXML files of records (the dataset where you want to add a classification, so for example the records that do not have a classification yet) as input and generates augmented MARCXML documents as output, seamlessly integrating the assigned classifications. Our primary objective is to demonstrate the practicality and adaptability of this method, with a particular emphasis on ensuring user-friendly interactions and transparent results. We made the workflow based on Python scripts because of its well-earned reputation for user-friendliness. Python’s syntax and readability make it particularly accessible, even for individuals with limited programming experience. This ease of use was a critical factor in our initial exploration as it allowed our team to quickly prototype and implement the scripts essential to our research without facing unnecessary complexities. It also aligned with our objective of creating a solution that could be easily adopted and adapted by a broad range of users. That is why we did not systematically assess or compare it against alternative programming languages in this particular context.
The Challenge of (Automated) Subject Indexing
Subject indexing, a crucial aspect of organizing information, has traditionally relied on manual methods to categorize content based on its subject matter. The conventional approach involves human indexers carefully reading and analyzing documents to assign relevant subject headings or terms. While this method has been the cornerstone of information organization for years, it comes with inherent limitations. Traditional subject indexing relies heavily on human expertise. The process demands skilled indexers who are knowledgeable in various domains. It can also be time-consuming and the dependence on human input affects the overall efficiency and accuracy of the indexing process. Moreover, as the volume of information grows exponentially, manually indexing a vast array of documents becomes increasingly impractical. The traditional approach struggles to scale effectively to handle the sheer magnitude of digital content generated daily. Recognising the limitations of traditional subject indexing, there is a growing need for a more streamlined and automated approach. An automated system can address these challenges and enhance the efficiency of information organization. First, automation can significantly accelerate the subject indexing process, allowing for the rapid categorization of large volumes of content. Secondly, automated systems help ensure a consistent and standardized application of subject headings. This minimizes subjective variations and ensures a higher level of accuracy across the indexed content. Thirdly, automated approaches are inherently scalable and well-suited for handling vast datasets. They can adapt to the increasing volume of information, providing a solution that is both efficient and sustainable. Lastly, maybe the biggest advantage is that automated subject indexing can leverage advancements in technology, such as natural language processing and machine learning, to improve the accuracy and relevance of automatically assigned subject terms. This integration enables a more sophisticated and adaptable indexing process.
The possibilities of Artificial Intelligence
Subject indexing has been a pioneering and widely adopted application of artificial intelligence (AI) in libraries, effectively streamlining the process of organizing and retrieving information [3]. However, its implementation is not without its challenges. Two primary approaches to subject indexing using AI can be identified: using a self-trained model or employing a large language model.
Training models for subject indexing can be a challenging and time-consuming endeavor. While the customization and potential accuracy offered by self-trained models are appealing, the process of constructing a comprehensive training dataset and effectively monitoring the model’s performance can be quite demanding. One of the primary difficulties lies in building a sufficiently large and representative training dataset. This dataset should encompass a wide range of subject terms and the corresponding texts they represent. Acquiring such a dataset can be a lengthy and labor-intensive process, often involving the manual extraction and compilation of data from various sources, such as library catalogs and online repositories. In our exploration of this option, we created a dataset using SPARQL queries to extract summaries from data.bnf.fr. This process entailed first identifying the subject terms we wished to include and then, for each term, locating at least ten associated summaries. This proved challenging, particularly for subject terms with a limited publication history or those specific to particular disciplines. Even after establishing a comprehensive training dataset, the task of monitoring and refining the self-trained model remained a continuous process. As the model encounters new documents to index, it may occasionally generate inaccurate, biased or misleading suggestions. Identifying the root cause of these misclassifications can be complex, as the model’s decision-making processes are often opaque and difficult to interpret. Without a clear understanding of why a particular term was suggested and not another, it becomes challenging to effectively refine the training dataset and improve the model’s performance. This lack of transparency can hinder the development of a truly reliable and consistent subject indexing tool.
In contrast, large language models like OpenAI’s ChatGPT or Google’s Bard offer a more automated and scalable approach to subject indexing. These models are trained on massive amounts of text data, enabling them to grasp the intricacies of human language and classify texts with greater accuracy. However, this versatility comes with the potential for inconsistency and a lack of control over the terminology used.
In summary, while both self-trained models and large language models hold promise for automating subject indexing in libraries, each approach presents unique challenges. Self-trained models offer greater customization and the potential for enhanced accuracy, but their training and monitoring can be demanding and opaque. Large language models provide scalability and automation but may lack consistency and standardized terminology. The choice between these approaches ultimately depends on the specific needs and priorities of the library or organization.
Words Matter: An Explanation of our Approach
To better understand how human catalogers determined the subject of a text, we observed their work processes. There are many things that determine a classification such as the author’s biography, or the publisher’s name (one publisher published only law books, for example), as well as the cover (children’s books or cookbooks are easily recognised by the cover), but of course also the back cover text. It was noteworthy that some catalogers indicated that they did not read that entire text in depth, but rather “scanned” the text, looking for key words of that text, to then determine the classification based on those key words. Human catalogers primarily rely on the presence of specific keywords and phrases within a text to infer its subject matter. This recognition inspired our approach to developing a system that places a central focus on the semantically significant words. In contrast to methods that often involve training sets with entire sentences, we opted for a distinctive system that centers around the extraction and analysis of specific words. Our system operates on the premise that these individual words serve as the building blocks for comprehending and categorizing content. We have devised a mechanism that suggests terms based on the contextual use of these linguistic units. This departure from sentence-level training sets to a more granular emphasis on words not only aligns with the observed practices of human catalogers but also presents a pragmatic solution. Our methodology prioritizes the semantically significant words, recognizing them as crucial elements in the intricate process of classification.
This approach, while intuitively appealing, is not without its limitations. It can be subjective and prone to errors, especially when dealing with complex or nuanced topics. For instance, a text about the history of the Middle Ages might explicitly mention “knights,” “castles,” “crusades,” and “counts,” leading a human cataloger to confidently assign the subject category “Middle Ages”. However, a more nuanced analysis might uncover subtler themes related to social structure, political dynamics, or cultural developments, requiring a more comprehensive approach. But our primary objective was to establish a general classification for the Belgian publications. For this purpose, a broad classification, such as “Middle Ages” would suffice. Furthermore, we sought a system that would limit its subject suggestions to the specific terms defined within our classification system for the Belgian Bibliography. The system should not propose any term outside of our national bibliography’s classification framework. Lastly, we also wanted a system that offers a more manageable and interpretable approach, allowing for greater control over the indexing process and enabling subject experts to refine and adapt the indexing rules as needed.
Our work aims to bridge the gap between the intuitive and practical approach of human catalogers and the more sophisticated capabilities of natural language processing (NLP) based methods. By combining these strengths, we hope to develop a practical and accurate solution for subject indexing in the context of Belgian languages (primarily Dutch and French), providing a valuable tool for improving the accessibility and discoverability of information in libraries and archives. While it may not achieve the same level of precision as self-trained models or large language models, it offers a more manageable and consistent approach for large-scale indexing projects.
Furthermore, our emphasis on the semantically significant words in the classification process aligns seamlessly with Python’s prowess in handling textual information. Python emerges as an ideal choice for automating tasks involving linguistic analysis, thanks to its extensive array of libraries and tools for data manipulation, analysis, and transformation. The intuitive syntax of Python substantially reduces the learning curve, enabling users, even those without extensive programming backgrounds, to swiftly develop scripts for diverse tasks. The active and robust Python community ensures continuous support and a wealth of documentation, facilitating troubleshooting and fostering collaboration [4]. Case in point, although we have backgrounds in history and art history, we successfully authored Python scripts that significantly aided us in improving and enriching our bibliographic and authority records. Consequently, it is unsurprising that, in our pursuit of an automated subject solution, we naturally turned our attention to Python.
In our pursuit of an optimal workflow, we established several criteria. First, the creation of classification had to be clear and verifiable. Equally significant was the ease of maintenance for this reference dataset. Thirdly, transparency in the output of terms was a key priority: we sought to understand the rationale behind terms presented and, in the event of inaccuracies, an easy path for correction. Accessibility was paramount, aiming for corrections to be effortlessly executed even by individuals with basic knowledge, opting for user-friendly tools such as Excel rather than more complex formats like XML or JSON. Lastly, the correctness of suggestions was imperative, ensuring that our workflow not only facilitated ease of use but also provided a tangible advantage in terms of accuracy. These criteria directed our developmental efforts, ensuring that the resulting workflow would be both effective and user-friendly.
The workflow is illustrated in the following diagram : from exported bibliographic records, used for the creation of the reference dataset, to the generation of enriched bibliographic records ready for import into our cataloging system. Our Library Management System (LMS) enables us to export records, apply batch modifications externally to the LMS, and subsequently re-import them to effect updates. This approach is adopted due to the inherent limitations of batch operations within our LMS, compelling us to optimize efficiency through external modifications.
Our workflow unfolds as follows: We initiate the process by exporting records from our Library Management System (LMS), each already assigned a classification. We export the pertinent information from these records, which is the title (245$a), remainder of the title (245$b), summary (520$a), and their corresponding classifications. Opting for a CSV export facilitates this operation, as we only require specific fields, and configuring an export profile tailored to these fields proves more manageable. Subsequently, our first Python script is executed to organize the texts by classification, with the associated words from the meaningful texts appended in the second column. The outcome is a CSV file presenting individual classifications paired with their respective meaningful words, forming our reference dataset. Afterwards, we further refined the reference dataset with batch ’replace all’ operations to eliminate less meaningful words and verbs.
Moving forward, we needed records awaiting classification, denoted as our input.xml. Our second Python script extracts the same crucial fields (245$a, 245$b, 520$a) and cross-references them with the words present in the reference dataset. Subsequently, it suggests a classification and appends it to the bibliographic record (084$a). The script also incorporates print statements, to provide insights into the considered words and elucidate the reasoning behind a particular term’s prominence. This information proves valuable in manual corrections of the reference dataset by adding or removing specific words associated with a category.
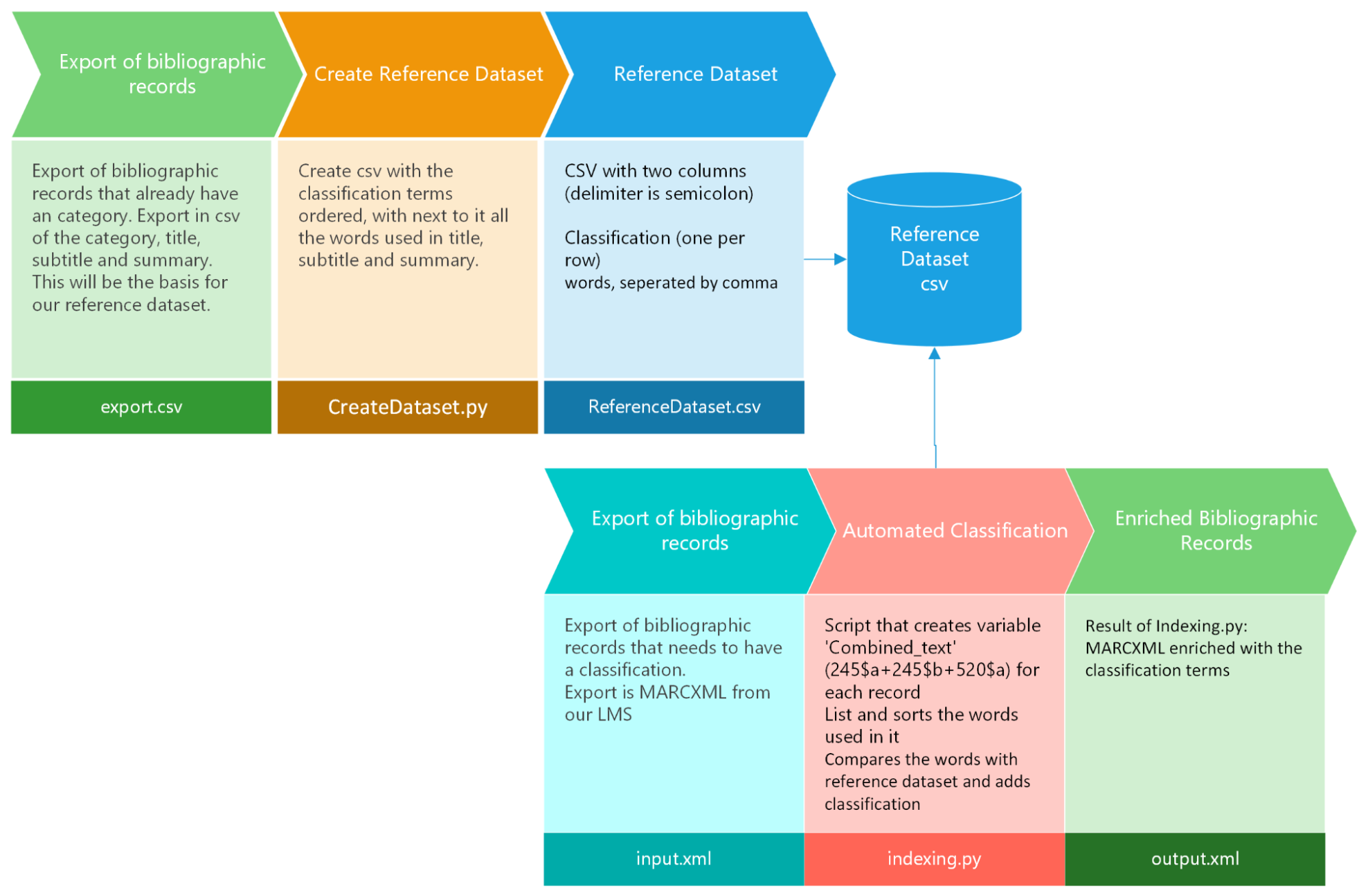
Figure 1. Representation
Let’s explore now the two specific scripts that played a pivotal role in establishing our automated subject indexing workflow.
First script: Creating the Reference Dataset
Creating the dataset is a pivotal step in our automated classification. Initially, we extract existing records from our database, focusing on those already classified. This extraction results in a CSV file, where classification terms (taken from our local classification system, that is stored in a local field 911$a were put in column ‘classifications’, while column ‘related terms’ includes relevant information such as title (245$a), subtitle (245$b), and summary (520$a). This script organises all records by classification term, and aggregates all associated words. In the process, stopwords (like articles) are deleted. This meticulous procedure culminates in the creation of our dataset, represented by the output file named “ReferenceDataset.csv.”
The code first imports the necessary libraries, including the Natural Language Toolkit (NLTK) [5], stopwords, and the csv library. It then downloads the NLTK stopwords corpus, which contains a set of common words that are often filtered out for text processing. This is only necessary the first time.
import nltk from nltk.corpus import stopwords import csv nltk.download(’stopwords’) # Only necessary the first time
Next, we make a set of stopwords in English, French and Dutch. The filter_stopwords() function takes a word as input and checks if it is in the set and thus a stopword. If it is, it will not be considered. If it is not a stopword, the function returns the word in lower caps.
stopwords = set(stopwords.words(’english’) + \
stopwords.words(’french’) + \
stopwords.words(’dutch’))
def filter_stopwords(word):
return word.lower() not in stopwords
The function extract_unique_words() receives a text as its input and dissects it into individual words by employing the split() method. This results in a word list, disregarding any punctuation or whitespace characters in the text and eliminating stopwords. The isalpha() method is applied to verify that each word solely consists of letters (a-z and A-Z, including letters with accents). Words containing non-alphabetic characters are discarded, a deliberate choice to minimize meaningless data. Consequently, words that are years or centuries are omitted, though it can be manually incorporated (e.g., as an associated term related to the ’history’ term). Finally, the function appends the unique words to a set and returns the list of these distinctive words.
def extract_unique_words(input_text):
words = input_text.split()
unique_words = set()
for word in words:
cleaned_word = word.strip(’.,’)
if filter_stopwords(cleaned_word) and (cleaned_word.isalpha()):
unique_words.add(cleaned_word.lower())
return list(unique_words)
Next, we define the names of input and output file.
input_file_name = ’export.csv’ output_file_name = ’ReferenceDataset.csv’
The code then creates an empty dictionary dict_classifications to store the extracted unique words for each classification. It iterates through the CSV data, reading each row as a list using next() to skip the header row. For each row, it extracts the classification and input text, calls the extract_unique_words() function to get a list of unique words, and checks if the classification exists in the dict_classifications dictionary. If the classification is not found, it adds a new entry to the dictionary with the classification and the list of unique words. If the classification exists, it checks if any of the unique words are already present in the dictionary’s value for that classification. For each unique word, if it’s not already present in the classification’s value, it appends it to the list. Finally, it iterates through the dict_classifications dictionary, creating a new row for each key-value pair and writing it to the output CSV file using csv.writer().
with open(input_file_name, ’r’, encoding=’utf-8’) as infile, \
open(output_file_name, ’w’, encoding=’utf-8’, newline=’’) as outfile:
ListReader = csv.reader(infile, delimiter=’;’)
ListWriter = csv.writer(outfile, delimiter=’;’)
dict_classifications = {}
next(ListReader)
for row in ListReader:
classification = row[0]
input_text = row[1]
list_result = extract_unique_words(input_text)
if classification not in dict_classifications:
dict_classifications.update({classification: list_result})
for i in list_result:
if i not in dict_classifications[classification]:
dict_classifications[classification].append(i)
else:
pass
for key, value in dict_classifications.items():
ListWriter.writerow([key, ’, ’.join(value)])
Once this script is executed, you’ll have a dataset ready for deployment in the subsequent script. However, we found it beneficial to perform some manual cleaning by deleting and eliminating extraneous words—those not essential or contributory to the classification (such as names, certain verbs, places, and occasionally dates (but some were kept because they might enhance the classification in categories like ’history’).
Table 1 : End result of script CreateDataset.py (after deleting non-significant words like author, monograph, book)
|
export.csv (existing records: classification; 245$a+245$b+520$a |
ReferenceDataset.csv (after script CreateDataset.py) |
|---|---|
|
Biology;The Selfish Gene. This book explores evolution though the lens of genes, suggesting they frive behaviors that maximize their own survival. History;A people’s history of the United States. This study offers a perspective on American history from marginalized voices and social movements. Biology;Sapiens. The author charts the history of Homo Sapiens, examining the biological factors that shaped human evolution. Sociology;Bowling Alone. This monograph explores the decline of social capital in modern society, highlighting the consequences of diminishing community engagement. History;The Silk Roads. This book traces the interconnected history of civilizations along the ancient trade routes, revealing cultural exchanges and influences. Sociology;The Sociological Imagination. This book encourages individuals to connect personal experiences with broader social structures and historical context. |
History;states, perspective, american, movements, marginalized, offers, people’s, study, united, voices, history, social, civilizations, influences, traces, interconnected, revealing, along, roads, trade, ancient, cultural, routes, exchanges, silk Biology;biological, examining, factors, homo, sapiens, human, charts, shaped, evolution, history Sociology;bowling, capital, alone, modern, society, social, diminishing, explores, engagement, highlighting, community, consequences, decline, broader, connect, personal, structures, experiences, encourages, imagination, individuals, sociological, historical, context |
In this example, we use the ‘textual’ terms (Biology, History, Sociology), but our model was trained using identifiers from our classification terms rather than the textual terms themselves. This approach enables us to seamlessly integrate enriched bibliographic records into our catalog, associating them directly with the accurate thesaurus term or topical name authority rather than relying solely on a text field containing the term.
Second script: Automated Indexing
The automated indexing script represents a crucial advancement in our subject indexing strategy, streamlining the intricate process and enhancing efficiency. At its core, this script aims to automate the assignment of a classification to bibliographic records based on their content.
Upon completing the ReferenceDataset.csv, the next step involves exporting the bibliographic records that needed a classification. We opted for a method that exports the entire bibliographic record, appending the classification in a designated field. In this article it adds a MARC21 field 084 (other classification number), but in our environment we use a local field 911, specifically created by our LMS provider to add a thesaurus. So we export bibliographic data from our lms (=‘input.xml’). This file is treated by the python script ‘indexing.py’. The result is an enriched MARCXML file that can be seamlessly re-imported into our LMS to update the existing records (=’output.xml’).
The script operates in two main phases. First, it analyzes the curated ReferenceDataset.csv, organizing records by classification term and compiling associated words. The script then employs natural language processing techniques to establish connections between subject terms and the content of the bibliographic records. It counts how many times each associated word is used in the combined text. It sorts the related terms based on that count and then adds the top three words as classification suggestions. Let’s delve into the key components of the script and explore its functionality:
1. Import libraries
The code first imports the necessary libraries and modules for the task, including CSV, xml.etree.ElementTree, and NLTK.
csv: To read and parse the CSV file containing suggested terms and associated words.xml.etree.ElementTree: To parse and modify the MARCXML files.NLTK: Here we use the tokenize function from the NLTK to perform NLP analysis on the text extracted from the MARCXML files.
import csv import xml.etree.ElementTree as ET from nltk.tokenize import word_tokenize
2. Define function to extract text
The getElementValue() function is used to extract text from our MARCXML file. It takes two arguments: an XML element and a separator string. The element can be a single element or a list of elements. The separator string is used to join the values of the elements. The function first checks if the element is None. If it is, then the function returns an empty string. If the element is not None, then the function checks if it is a list. If it is, then the function iterates over the list and appends the text of each element to a list. The function then joins the values of the list with the separator string and returns the joined string. If the element is not a list, then the function checks if it has a text attribute. If it does, then the function returns the value of the text attribute. If the element has neither of these properties, then the function returns an empty string.
def getElementValue(elem, sep=’;’):
if elem is not None:
if isinstance(elem, list):
valueList = list()
for e in elem:
if hasattr(e, ’text’):
if e.text is not None:
valueList.append(e.text)
return ’;’.join(valueList)
else:
if hasattr(elem, ’text’):
return elem.text
return ’’
3. Parse CSV file
Next, the code reads the ReferenceDataset.csv file using the csv library and creates a dictionary term_associations using the CSV data (the ReferenceDataset.csv). Each key represents a subject term and the corresponding value is a list of associated words.
with open(’ReferenceDataset.csv’, newline=’’, encoding=’utf-8’) as csvfile:
csv_reader = csv.reader(csvfile, delimiter=’;’)
term_associations = {rows[0]: rows[1].split(’, ’) for rows in csv_reader}
4. Parse MARCXML File and extract text
The code parses the input.xml file using the ET library and extracts the root collection node from the parsed XML document. It also defines the namespace for the MARCXML. Next, the code iterates through each record in the collection node. For each record, it creates an empty string combined_text to store the combined text from specific data fields. It processes each record and extracts the title (245$a), remainder of title proper (245$b), and summary note (520$a) from our MARC21 record, using the pre-defined getElementValue function and XPath expressions. It appends the values of MARC21 fields 245$a, 245$b and 520$a to the combined_text string. It then prints the combined text (for debugging if necessary).
ET.register_namespace(’’, ’http://www.loc.gov/MARC21/slim’)
tree = ET.parse(’input.xml’)
collection = tree.getroot()
all_ns = {’marc’: ’http://www.loc.gov/MARC21/slim’}
for record in collection:
combined_text = ""
Field245a = getElementValue(record.find(\
’./marc:datafield[@tag="245"]/marc:subfield[@code="a"]’, all_ns))
Field245b = getElementValue(record.find(\
’./marc:datafield[@tag="245"]/marc:subfield[@code="b"]’, all_ns))
Field520a = getElementValue(record.find(\
’./marc:datafield[@tag="520"]/marc:subfield[@code="a"]’, all_ns))
combined_text = Field245a + " " + Field245b + " " + Field520a
print("Combined Text:", combined_text)
5. NLTK Word Tokenization
The next part of the script focuses on tokenizing and filtering the combined text for further analysis. The word_tokenize function from NLTK is used to tokenize the combined_text into individual words. This function considers punctuation, contractions, and other special characters as separate tokens. Then, the script iterates through the tokens and performs two filtering steps:
- Lowercase Conversion: Converts all tokens to lowercase using a list comprehension. This helps normalize case differences and prepare for further analysis.
- Alphanumeric Check: Keeps only tokens containing alphanumeric characters (letters and numbers) using the isalnum() method. This removes punctuation, symbols, and other non-word elements.
A variable count is initialized to 0, for later use in counting occurrences of specific words or other analysis tasks. Overall, this part of the script breaks down the text into individual words, converting them to lowercase, and filtering out non-alphanumeric characters.
# NLTK tokenization and analysis for French texts tokens = word_tokenize(combined_text) filtered_tokens = [word.lower() for word in tokens if word.isalnum()] count = 0
6. Counting the occurrences of words
This part of the script focuses on counting the occurrences of associated words in the filtered combined text. This part analyzes the filtered_tokens by counting how often each associated word (from the term_associations created in step 3) appears in it. It creates a word_counts dictionary to store these occurrences. This provides insights into the presence of relevant terms based on their associated words.
For each associated word, it counts the number of occurrences in the text using the sum() function. Based on the word_counts, the terms are sorted and saved in the sorted_terms dictionary. In the provided script, the occurrences of associated words in the combined text are counted individually. This means that if an associated word appears more than twice in the combined text, it will be counted as multiple matches. This is done to ensure that all instances of the associated words are considered when determining the relevance of a subject term. The script maintains a word_counts dictionary to track the occurrences of each associated word. This dictionary is used to sort the term associations based on the overall frequency of their associated words. As a result, subject terms with more associated words and higher occurrence counts are considered more relevant. The sorted_terms are limited to the top three suggestions.
word_counts = {word: sum(1 for token in filtered_tokens \
if token.lower() == word)
for word in set(word for words in term_associations.values()
for word in words)}
sorted_terms = sorted(term_associations.items(), \
key=lambda x: \
sum(word_counts[word] for word in x[1]), reverse=True)[:3]
7. Extract Subject Terms
Next, we initialize an empty list subject_terms to store the potential subject terms. For each term and its associated words, it creates a list matched_words containing only associated words that appear in the filtered_tokens. In other words: the matched_words list is populated with words from the associated_words list that appear in the text. If matched_words is not empty:
- It counts the number of unique matched words using the
len()function. - It adds the term to the
subject_termslist. -
It prints information about the matched term (useful for analysis and monitoring):
- The term itself.
- The list of matched words.
- The total number of occurrences (summed from
word_counts)./li> - The number of unique matched words.
- A newline for formatting.
It prints the final list of extracted subject terms (subject_terms), followed by a separator (—-) for clarity.
subject_terms = []
for term, associated_words in sorted_terms:
matched_words = [word for word in associated_words \
if any(token.lower() == word \
for token in filtered_tokens)]
if matched_words:
unique_word_count = len(set(matched_words))
subject_terms.append(term)
print(f"Matched Term: {term}")
print(f"Matched words: {matched_words}")
print(f"Total occurrences: {sum(word_counts[word] for word in associated_words)}")
print(f"Total unique word count: {unique_word_count}")
print()
print("Final subject terms:", subject_terms)
print("----")
8. Add 084subfields
The next part adds the subject term in MARC21 084 subfields to the MARCXML records for the top three extracted subject terms based on the analysis done previously.
for term in subject_terms:
datafield_084 = ET.SubElement(record, ’datafield’, tag="084", \
ind1=" ", ind2=" ")
subfield_a = ET.SubElement(datafield_084, ’subfield’, code="a")
subfield_a.text = term
subfield_7 = ET.SubElement(datafield_084, 'subfield', code="7")
subfield_7.text = 'automatically generated'
9. Save modified XML
Finally, the code saves the modified MARCXML record to the file output.xml.
tree.write(’output.xml’, xml_declaration=True, encoding=’utf-8’, method="xml")
Case Study
Let’s illustrate the functionality using an example. Consider a bibliographic record with the following fields:
<datafield tag="245" ind1="0" ind2="0">
<subfield code="a">Societal Shifts</subfield>
<subfield code="b">A New approach</subfield>
</datafield>
<datafield tag="520" ind1=" " ind2=" ">
<subfield code="a">
This book delves into the intricate fabric of contemporary communities.
Drawing on a rich tapestry of sociological theories and empirical
studies, the book examines key themes such as the impact of
globalization, technological advancements and shifting cultural norms
on the structure of societies. Through insightful analyses, the author
dissects the evolving nature of interpersonal relationships, the
emergence of new social inequalities, and the role of institutions
in navigating these transformations. This sociological exploration
goes beyond observation, offering readers a profound understanding
of the complex web of relationships that define our modern social
landscapes
</subfield>
</datafield>
If we launch the script, we get the following output:
- Combined Text: Societal Shifts A new approach This book delves into the intricate fabric of contemporary communities. Drawing on a rich tapestry of sociological theories and empirical studies, the book examines key themes such as the impact of globalization, technological advancements and shifting cultural norms on the structure of societies. Through insightful analyses, the author dissects the evolving nature of interpersonal relationships, the emergence of new social inequalities, and the role of institutions in navigating these transformations. This sociological exploration goes beyond observation, offering readers a profound understanding of the complex web of relationships that define our modern social landscapes
-
Words (sorted by occurrence, and those with 0 occurrence excluded):
-
sociological: 2
- one of the associated words related to classification ‘Sociology’, counted as 2 because the word appears two times in the combined text.
-
social: 2
- one of the associated words related to classification ‘Sociology’, counted as 2 because the word appears two times in the combined text.
- also one of the associated words related to classification ‘History’, counted as 2 because the word appears two times in the combined text.
-
modern: 1
- one of the associated words related to classification ‘Sociology’
-
personal: 1
- one of the associated words related to classification ‘Sociology’
-
cultural: 1
- one of the associated words related to classification ‘History’
So the result of this small example is
Sociology: 5 occurrences of 3 matched words
History: 3 occurrences of 2 matched words
Biology: 0 matches -
sociological: 2
- Final subject terms: [’Sociology’, ’History’]. The proposed terms are sorted by number of occurrences, as it is considered as best fitting. So that is why Sociology is presented first.
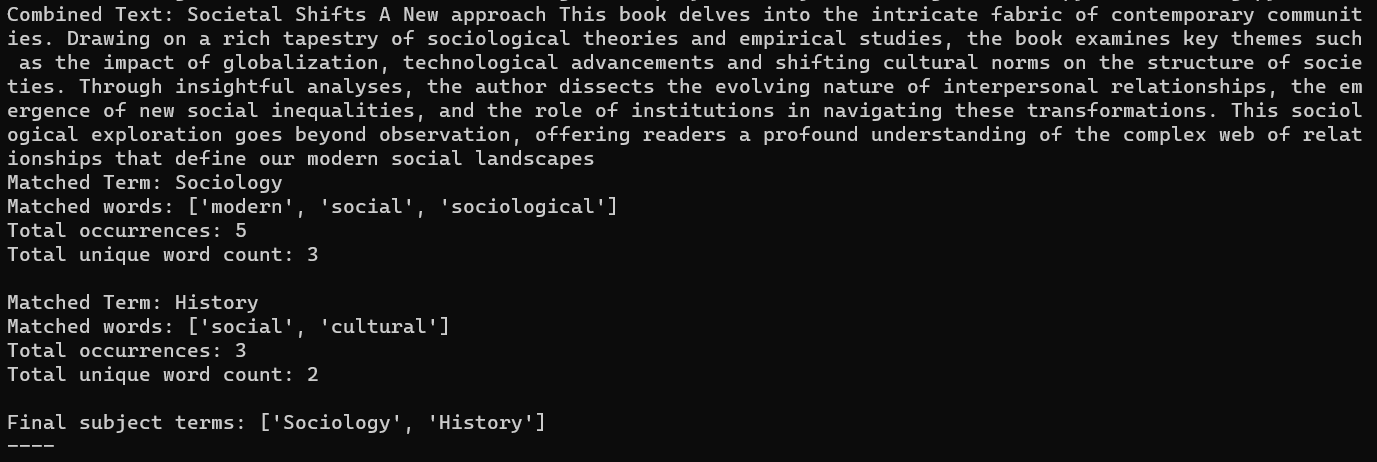
Figure 2. Outcome test
Certainly, the effectiveness of matching improves with a greater number of associated words related to the terms. If the results prove satisfactory, we have the flexibility to adjust and potentially limit the matching to just one term instead of three. One notable advantage lies in the ease with which we can improve our ReferenceDataset.csv. This can be achieved effortlessly through manual additions or by incorporating words from other bibliographic records. Notably, any changes or additions to associated words take immediate effect without the need for retraining. Unlike systems relying on NLP that necessitates training based on new words, our approach involves a straightforward comparison of utilized words. This inherent flexibility ensures the system is easily adaptable and maintainable, accommodating dynamic adjustments to suit evolving requirements.
Conclusion
In conclusion, our exploration of simplifying subject indexing at KBR, the National Library of Belgium, has unveiled a promising Python-powered approach. We began by acknowledging the necessity of efficiently classifying our extensive collection through legal deposit. Recognizing the growing need for a streamlined and automated approach, we explored two primary AI-based strategies: self-trained models like Annif and large language models such as OpenAI’s ChatGPT of Google’s Bard. While each approach carries its own advantages and challenges, the choice ultimately depends on the specific needs and resources of each library. Despite the advancements in AI-based subject indexing, we sought a third alternative. Our focus shifted to Python as a solution, emphasizing its power and simplicity in automating tasks across diverse domains.
Introducing two specific scripts—CreateDataset.py and indexing.py—we demonstrated the creation of a reference dataset and the subsequent automated subject indexing workflow. The creation of the dataset included taking the values of some fields, filtering out extraneous words, but also some manual cleaning was necessary. The indexing script automated the assignment of classifications to bibliographic records based on their content, using natural language processing and associating words from a curated dataset. Our case study exemplified the script’s functionality, showing the extraction of subject terms based on the associated words. It ends with adding the classification to the MARCXML records. The script’s flexibility allowed for customization, limiting the number of suggested terms (and control about which terms/identifiers can be suggested and thus imported) and offering immediate adaptability to changes in the reference dataset.
We believe this method proves particularly useful in situations where there is limited text or summary available, such as when dealing solely with a title and/or subtitle because the approach relies on keywords (the ’associated words’ linked to each term). For instance, it can assist in classifying old and rare books, which often lack detailed summaries but have meaningful titles with significant words like ’biblia’, ’necrologia’, ’vita’ that can help to classify the works.
An additional benefit is the clarity provided by the print statements, allowing a thorough analysis of why a particular classification term was suggested and identifying the words it matched. Rapid correction of associated words can follow, enhancing the overall outcome. The user-friendly nature of this approach facilitates a clear comprehension of its workings. In our view, it strikes a good balance between automation and manual management of the reference dataset, contributing to improved results.
In essence, our two scripts in Python proved to be a potent solution for automation, providing unparalleled flexibility and simplicity in handling data processing tasks. The ease of use and adaptability of the approach make it a compelling choice for libraries working on subject indexing projects. We encourage readers to explore the possibilities of Python in their own endeavors, leveraging its rich ecosystem and community support to enhance efficiency and maintainability in information organization.
Notes
[1] https://annif.org/. See also Osma Suominen, Ilkka Koskenniemi, Annif Analyzer Shootout: Comparing text lemmatization methods for automated subject indexing, Code4Lib Journal, 2022-08-29 (issue 54).
[2] https://learn.microsoft.com/en-us/ai-builder/text-classification-overview
[3] Already two years ago, the Cataloging & Classification Quarterly attributed a whole issue (issue 8, 20221, volume 59) to the topic of ‘Artificial Intelligence and Automated Processes for Subject Access.
[4] Recently, a clear overview of the status and advantages of using Python was published in Code4Lib Journal: Collins M, Song X, Schon S. 2023. The use of Python to Support Technical Services Work in Academic Libraries. Code4Lib Journal [Internet]. 58:2023-12-04. Available from:https://journal.code4lib.org/articles/17701
[5] See https://github.com/nltk/nltk and https://www.nltk.org/
About the authors
Hannes Lowagie (he/him) is head of the Agency for Bibliographic Information in KBR, the national library of Belgium. He is currently working on the transition to RDA, Linked Open Data and the use of AI-technologies in the cataloguing process. He has a PhD in (Medieval) History (University of Ghent, 2012).
Julie Van Woensel (she/her) is a metadata librarian at KBR, the Royal Library of Belgium, where she works on improving bibliographic and authority records and supporting the transition towards RDA and linked data.


Subscribe to comments: For this article | For all articles
Leave a Reply